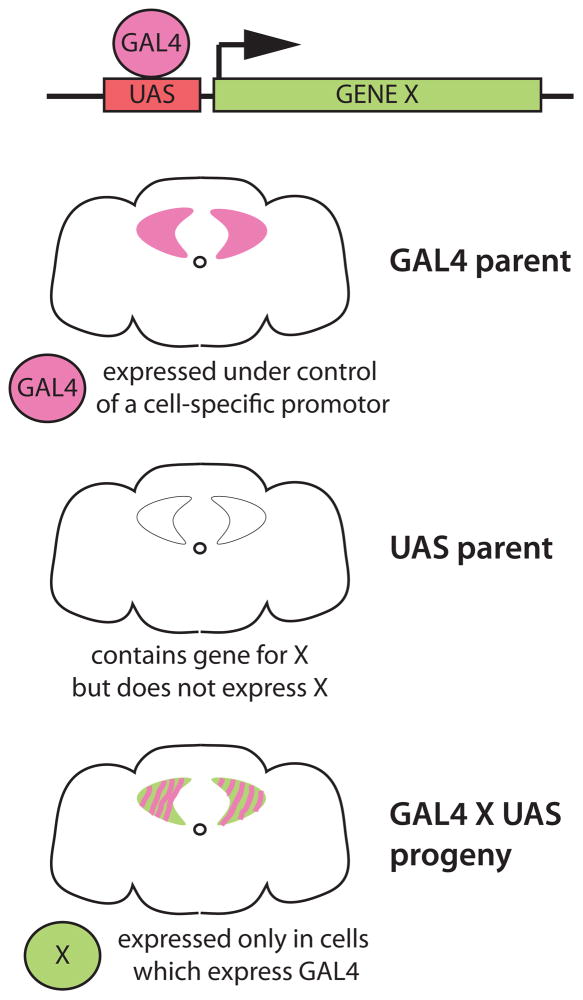
Figure 1.
The GAL4/UAS system for cell-specific transgene expression [8]. The yeast transcriptional activator GAL4 is typically expressed as a transgene under control of local DNA elements by either random insertion of its cDNA or by homologous recombination of its cDNA into a defined locus (e.g. the fru gene [38]), to capture that gene’s expression pattern. GAL4 can also be expressed under control of cloned promoter sequences (e.g. the pdf promoter which captures part of the clock circuit [18]). GAL4 lines are crossed to effector lines containing a transgene with a GAL4-binding UAS (upstream activation sequence) repeat upstream of the coding sequence of the gene of interest (in this example, X). Expression of X is dependent on GAL4, and only occurs in the progeny of the cross in cells that express GAL4. The Drosophila community has generated thousands of GAL4 lines, allowing specific genetic access to most of the nervous system. There are also variants of GAL4 that can further restrict expression patterns temporally and spatially, as well as several independent binary systems that can be used along side GAL4/UAS. A complete guide to these systems and the effector tools that can be used with them is provided in [9].