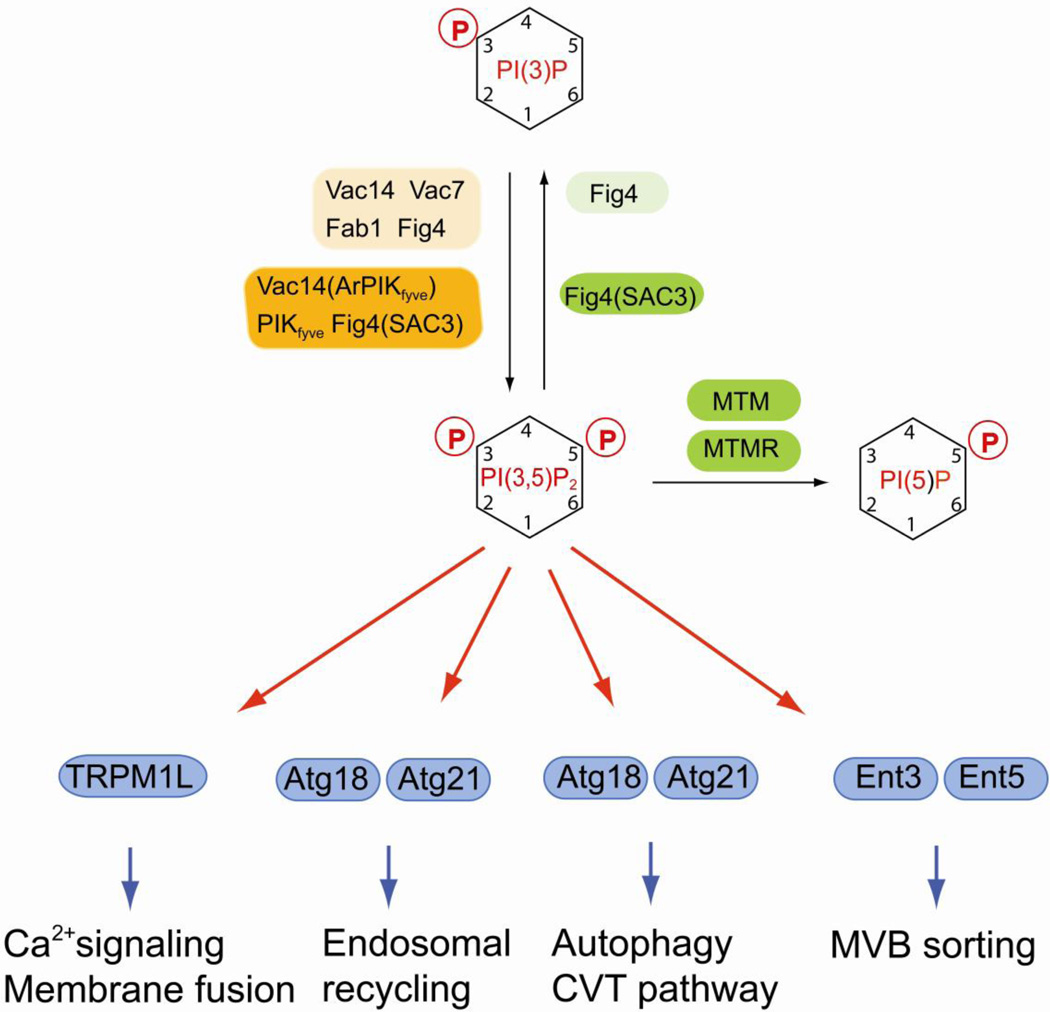
Fig. 2.
Regulation of PI(3,5)P2 and its function in late endosomal dynamics and in autophagy. PI(3,5)P is generated by the FYVE domain-containing lipid kinase Fab1. Synthesis and turnover of PI(3,5)P2 are tightly coupled. Yeast Fab1 is only active when it is in a complex with the scaffold protein Vac14 that also interacts with Vac7 and with the PI(3,5)P2 phosphatase Fig4. A similar complex is also found in mammals containing the lipid kinase PIKfyve, the scaffold protein Vac14 (also termed ArPIKfyve) and the lipid phosphatase Fig4 (also termed Sac3). PI(3,5)P2 plays multiple roles in late endosomal dynamics, autophagy, MVB sorting and Ca2+ mediated membrane fusion. Mammalian lipid kinases or kinase complexes are marked by orange boxes, lipid phosphatases are in green boxes. Alternative names for certain enzymes are in parentheses. Yeast homologs are indicated by lighter shades of the same colors. Effector proteins are in blue boxes.