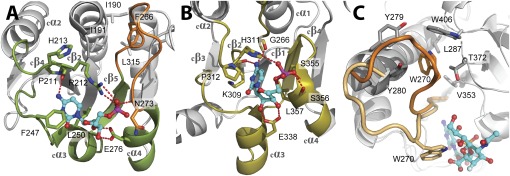
Fig. 4.
Comparison of the donor-substrate binding sites of WaaAAAE (A) and a sialyltransferase of the GT-B superfamily [e. g., PM0188 (19)]. (B). Equivalent protein regions involved in the binding of the CMP moiety of the donor substrate are highlighted in green (WaaAAAE) or olive (PM0188). In both structures, an α/β/α motif, overlapping with the α/β/α motif of MurG (16), is involved in CMP binding. Hydrogen bonds are depicted as red dotted lines, and residues involved in CMP binding are shown as sticks. Note that the positions of hydrophobic residues sandwiching the cytosine base (F247/L250 for WaaAAAE and P312/L357 in PM0188) and the glutamate residue contacting the ribose hydroxyls (E276 in WaaAAAE and E338 in PM0188) vary within the equivalent protein regions. The HP motif of PM0188 (H311/P312) overlaps with the highly conserved 211PRH213 triplet of WaaAAAE. The putative Kdo-binding loop of WaaAAAE in A is highlighted in orange. For clarity, the 3F-Neu5Ac moiety of the donor substrate in B has been omitted. (C) Conformational changes upon donor-substrate binding as evident from superimposed structures of PM0188 (23). In the apo form, W270 (orange) is part of a hydrophobic cluster. Binding of the donor-substrate analog CMP-3F-Neu5Ac brings W270 (beige) into contact with the 3F-Neu5Ac moiety of the donor substrate. For clarity, the perspective of C is different from B.