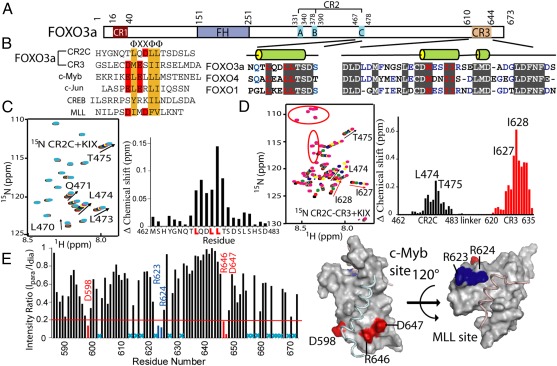
Fig. 1.
Characterization of interactions between FOXO3a and the CBP KIX domain. (A) Domain architecture of FOXO3a, indicating the FH domain and three conserved regions (CR1–3). Sequence alignments of CR2C and CR3 from FOXO proteins are shown with the predicted secondary structure. (B) Sequence alignment of KIX-interacting peptides. The hydrophobic residues of the “ΦXXΦΦ” motif are colored orange, and negatively charged residues are colored red. (C) NMR titration of 15N-labeled CR2C (0.2 mM) with KIX. (Left) HSQC of 15N-labeled CR2C (black) overlaid with spectra collected during titration with KIX: 0.1 (red), 0.35 (green), 0.85 (blue), 1.35 (yellow), 1.85 (pink), and 4.75-fold (cyan). Perturbed resonances are labeled. (Right) Normalized chemical shift change of each CR2C residue on addition of 4.75-fold excess KIX. The “LXXLL” motif is highlighted. (D) NMR titration of 15N-labeled CR2C-CR3 (0.16 mM) with KIX. (Left) HSQC of 15N-labeled CR2C-CR3 (black) overlaid with spectra collected during titration with KIX: 0.25 (red), 0.75 (green), 1.75 (blue), 3.25 (yellow), and 4.75-fold (pink). Perturbed resonances are labeled. Red circles indicate Gly and Ser residues in the engineered linker. (Right) Normalized chemical shift change of each CR2C-CR3 residue on addition of 4.75-fold excess KIX. CR2C and CR3 residues are plotted in black and red, respectively, and the most perturbed residues are labeled. (E) Probing CR2C-CR3 binding to KIX by PRE. (Left) Peak intensity ratio (Ipara /Idia) of each KIX residue in the presence of CR2C-CR3 peptide tagged with Mn2+ (para) vs. Ca2+ (dia). Cyan X's indicate residues for which this ratio cannot be determined because of overlapped peaks. Residues exhibiting large broadening (ratio <0.2, red line) are labeled. (Right) Broadened residues mapped on the ternary KIX–c-Myb–MLL complex (PDB ID code 2AGH). The c-Myb and MLL sites are indicated, and broadened residues in each site are colored red and blue, respectively. All structure figures were prepared using PyMol (The PyMOL Molecular Graphics System, Schrödinger, LLC).