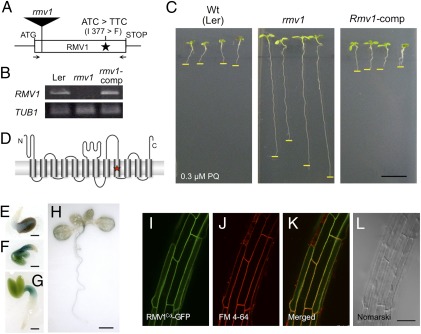
Fig. 2.
Characterization of RMV1. (A) Schematic representation of the structure of RMV1. The position of nucleotides and amino acid residues that differ between Ler-1 and Nos-d is indicated. The position of the T-DNA insertion of rmv1 (GT_3_3436) is shown as a filled rectangle. Arrows denote primers used for RT-PCR. (B) RMV1 transcript levels in wild-type (Ler-1), rmv1, and complementation transgenic lines (rmv1-comp). RMV1 transcripts were detected by RT-PCR using total RNA extracted from 2-d-old seedlings. (C) PQ sensitivity of the wild-type (Ler-1), rmv1, and complementation transgenic lines (rmv1-comp). (D) Schematic view of the topology of RMV1, as predicted by ConPred II (34). Transmembrane domains are indicated as cylinders. The star denotes the position of amino acid residues that differ between the RMV1 proteins derived from Ler-1 and Nos-d. (E–H) RMV1 promoter-GUS reporter gene expression in transgenic plants (E–H: 1, 2, 3, and 10 d after germination, respectively). (I–L) Subcellular localization of the RMV1c-GFP fusion protein. Roots of Arabidopsis stably transformed with RMV1Col-GFP and stained with FM4-64. (I) GFP filter. (J) FM4-64 filter. (K) merged. (L) Nomarski images. [Scale bars: 1 cm (C); 1 mm (E–G); 5 mm (H); 50 μm (L).]