Fig. 2.
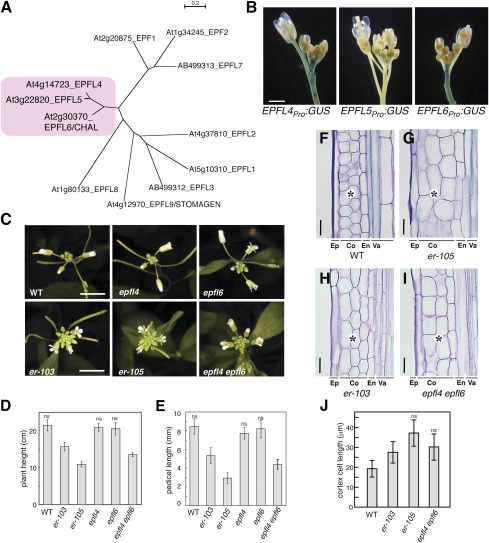
EPFL4 and EPFL6 redundantly control inflorescence growth in a manner similar to ER. (A) Molecular phylogeny of EPFL family members. EPFL4, EPFL5, and EPFL6 cluster together. Shown is an unrooted neighbor-joining tree of C-terminal end amino acid sequence encompassing the predicted mature EPF (MEPF) domain. Branch lengths are scaled to the number of amino acid changes indicated on the scale bar. MEPF sequence alignment is shown in Fig. S8. (B) Promoter activities of EPFL4, EPFL5, and EPFL6 in inflorescence. (Scale bar: 1 mm.) (C) epfl4 epfl6/chal-2 confers compact inflorescence nearly identical to that of er. Shown are inflorescence tops of 5-wk-old Arabidopsis of WT, epfl4, epfl6, er-103 (intermediate allele), er-105 (null allele), and epfl4 epfl6/chal-2. (Scale bars: 1 cm.) (D and E) Morphometric analysis of plant height (D) and pedicel length (E) from each genotype. 5-wk-old plants were measured for plant height (n = 9), and 6-wk-old mature pedicels (n = 46, from 6 plants) were measured. Bars indicate mean values; error bars indicate SD. ns, not significantly different and grouped together (Tukey's HSD test). (F–I) Longitudinal sections of mature pedicels from WT (F), er-105 (G), er-103 (H), and epfl4 epfl6/chal-2 (I). Ep, epidermis; Co, cortex; En, endodermis; Va, vasculature. epfl4 epfl6 has large cortex cells similar to er (asterisks). (Scale bars: 25 μm.) (J) Quantitative analysis of cortex cell length (n = 30). Bars indicate mean values; error bars indicate SD. ns, not significantly different and grouped together (Tukey's HSD test). The images in each composite were obtained under the same magnification.