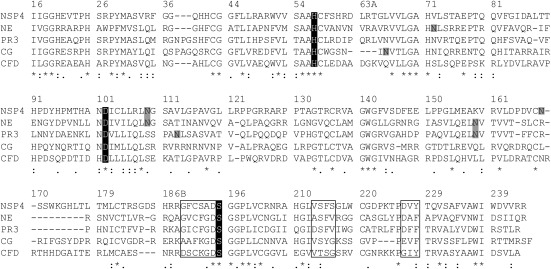
Fig. 1.
Sequence comparison between all four catalytically active human neutrophil serine proteases and complement factor D. Topologically aligned amino acid residues (single letter code) are numbered according to chymotrypsinogen. The catalytic triad of His57, Asp102, and Ser195 is highlighted in black. NSP4 carries two N-linked glycosylation sites marked in gray. Residues that shape the substrate binding pocket in NE are boxed (189–195, 213–216, 226–228) for all proteases and are similar between NSP4, NE, and PR3. An asterisk (*) below the alignments indicates positions which agree completely; a colon (:) or a period (.) indicate conservation of residues with strong or weak similarities (scoring > 0.5 or = < 0.5 in the Gonnet PAM 250 matrix), respectively.