Fig. 5.
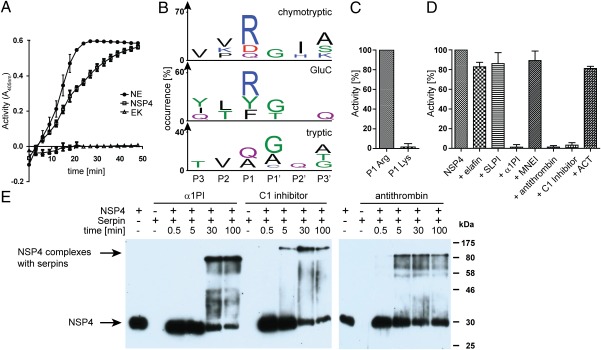
NSP4 preferably cleaves after arginine residues and is inhibited by α1PI, C1 inhibitor, and antithrombin. (A) After removing the propeptide extension with enterokinase (EK), proteolytic activity of recombinant NSP4 was measured using the thiobenzyl ester substrate Boc-Ala-Pro-Nva-4-chloro-SBzl and DTNB. NSP4 was used at a concentration of 3 × 10−6 M. NE and the convertase EK served as positive and negative controls at concentrations of 1 × 10−7 M and 3 × 10−2 U/μL, respectively. Substrate cleavage was determined by absorbance measurements at 405 nm. Data are results from three experiments, ± SD. (B) PICS specificity profile for NSP4 with chymotrypsin, GluC, and trypsin libraries generated from E. coli. Sequence logos were generated with IceLogo (26). (C) Enzymatic activity of 0.25 μM NSP4 was measured using AMC substrates with either Arg or Lys in P1 [H-Tyr-Arg-Phe-(Arg/Lys)-AMC] at 1 mM substrate concentration. Percent activity was calculated by setting the mean activity of NSP4 with P1 Arg to 100%. Data are the mean from three independent experiments (n = 3, ± SD). (D) 0.25 μM NSP4 was preincubated with 10 μM elafin, 10 μM SLPI, 25 μM α1PI, 3.3 μM MNEI with 1 mM DTT, 5.6 μM antithrombin with 300 U/mL heparin, 2.5 μM C1 inhibitor, or 5.6 μM ACT. Proteolytic activity was determined using H-Tyr-Arg-Phe-Arg-AMC. Percent activity was calculated by setting the mean activity of NSP4 without inhibitor to 100%. Data are the mean from three independent experiments (n = 3, ± SD), except for MNEI and ACT (n = 2, ± SD). (E) 0.8 μM NSP4 was incubated at 37 °C with 40 μM α1PI, 4 μM C1 Inhibitor or 5 μM antithrombin with 300 U/mL heparin and analyzed after different timepoints by Western blotting with anti NSP4 mAbs. For comparisons, NSP4 and the serpins alone were treated similarly.