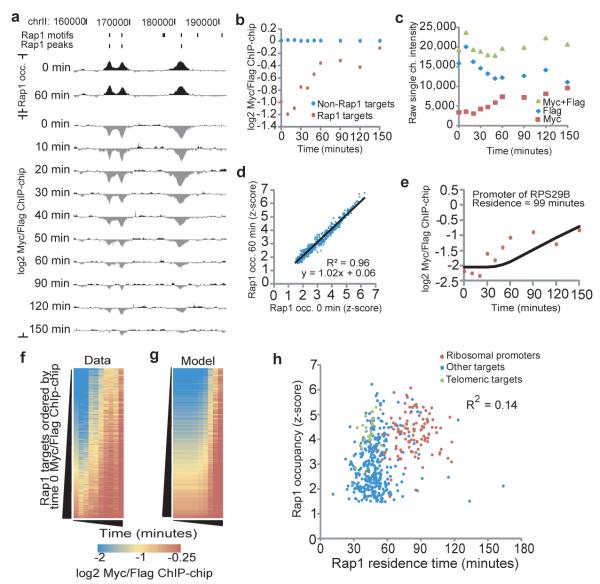
Figure 2. Rap1-bound sites exhibit distinct replacement dynamics.
(a) A Rap1 turnover experiment over a 30-kb region of chromosome II. Rap1 motifs and peaks are indicated. (b) Average log2 Myc/Flag values for all Rap1 targets (red) increase relative to non-Rap1 targets (blue). (c) Rap1-Myc competes with Rap1-Flag for binding. Average single channel intensity for Rap1-Myc and Rap1-Flag for a single probe (id:CHR15FS000978891) in the promoter of TYE7/YOR344C shows the increase in Rap1-Myc is coincident with the loss of Rap1-Flag. (d) Total Rap1 occupancy does not change during the time-course. Average total Rap1 occupancy (log2 Rap1 IP (y-300)/input z-score) at Rap1 targets at time 0 versus 60 minutes is plotted. (e) Average log2 Myc/Flag values for the promoter of ribosomal protein gene RPL29B (red points). The model fit for the residence time parameter that best fits this data is shown (black line). (f) Colorimetric representation of log2 Myc/Flag values for all 465 Rap1 targets, sorted by the initial (normalized) log2 Myc/Flag value. (g) For each site in (f), the log2 Myc/Flag value predicted by our residence model based on the calculated residence time. (h) Rap1 occupancy (time 0 z-score) vs. Rap1 residence for 465 Rap1 targets (R2= 0.14, 0.37 spearman rank correlation).