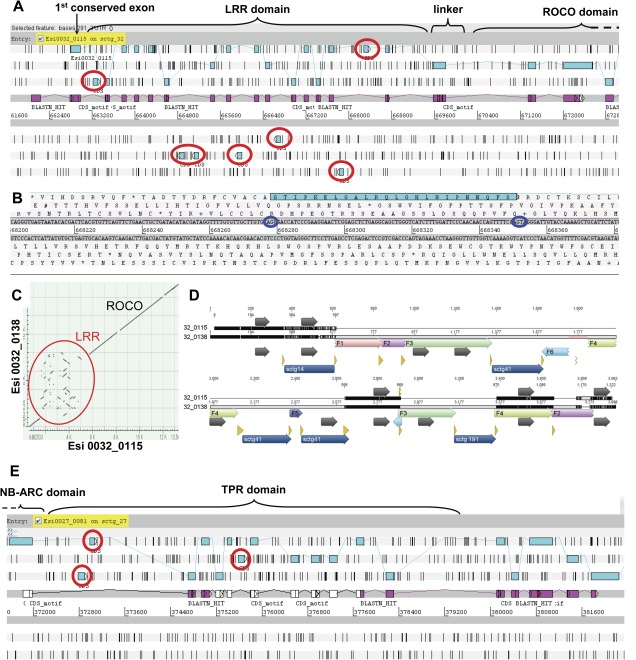
FIG. 2.
The genomic organization of Ectocarpus ROCO and NB-ARC–TPR genes reveals intense shuffling of LRR (respectively TPR)-encoding exons. (A) Genomic organization of the Esi0032_0115 ROCO locus. The predicted protein sequence (joined blue rectangles) is supported throughout by ESTs (mapped in pink on the coding strand). Additional noncoding LRR exons in the intronic regions appear as blue rectangles circled in red. (B) Zoomed view of an intact noncoding type I LRR exon, with its conserved splicing sites circled in dark blue. (C) Nucleotide identity dot matrix between the two recently duplicated ROCO loci Esi0032_0115 and Esi0032_0138. Both gene sequences are almost identical, except for their LRR domain that appears highly recombined. (D) Alignment of the genomic sequences of Esi0032_0115 and Esi0032_0138, along the first 3 kb of their LRR domain. Highly conserved regions between the two genes are in black, unconserved regions are in light gray. LRR exons (on the coding strand only) are depicted with dark gray arrows. The pastel boxes represent homologous, locally rearranged sequence fragments (numbered F1–F6, originating from intragenic recombination), whereas dark blue boxes at the bottom represent sequences inserted from other genome scaffolds (sctg 14, sctg 41, and sctg 191, originating from intergenic recombinations). Recombination sites are highlighted with the yellow arrows. The pale red area around position 1777 in the Esi0032_0138 sequence corresponds to a short interruption in the genome assembly. (E) Genomic organization of the TPR domain of the Esi0027_0081 NB-ARC locus. The predicted protein sequence is depicted in blue, partially supported by ESTs (mapped in pink on the coding strand). Additional noncoding TPR exons (as judged by the absence of splicing sites, the presence of a frame shift, or of a stop codon) in the intronic regions appear as blue rectangles circled in red.