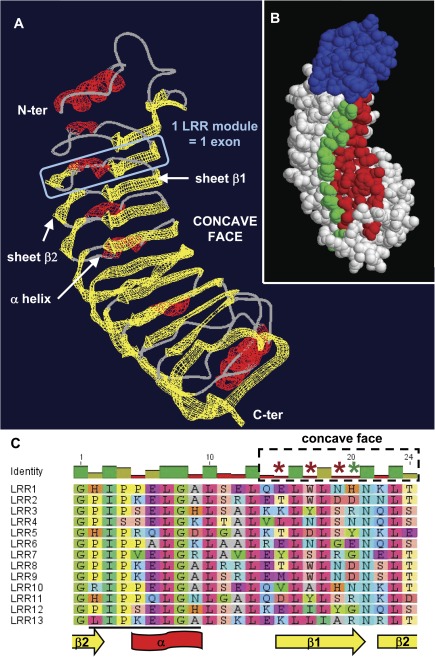
FIG. 4.
Positive selection acts on solvent-exposed and potentially ligand-binding residues of the LRR domains. (A) Homology-based 3D model of Esi0032_0115 N-terminus and LRR domain. The designation of β−β−α structures follows (Di Matteo et al. 2003). (B) Corresponding surface view of the polypeptide, with the potential location of positively evolving sites in type I LRR exons highlighted. Sites shown in red are those (15, 17, 19) where model M8 revealed posterior probabilities greater than 0.99, while site 20 (in green) was supported by a posterior probability of 0.88. All sites are predicted to be located on the concave, probably ligand-binding face of the domain. The N-terminal region is depicted of the protein in blue. (C) Alignment of the 13 exons composing the LRR domain, showing the conserved structural residues interspersed with variable amino acids. Asterisks highlight the four amino acid positions subject to positive selection, with the same color code as in (B). The region targeted by the esi_mir_cand_5 miRNA family (Cock et al. 2010) is underlined in black.