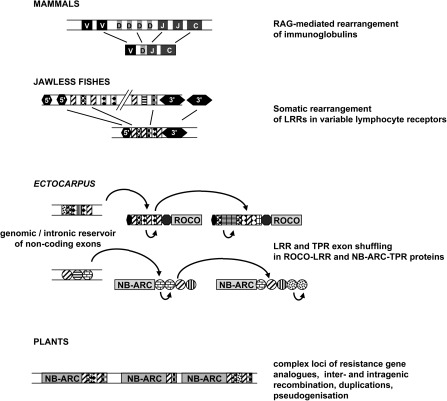
FIG. 5.
Exon shuffling in the Ectocarpus ROCO and NB-ARC proteins could underpin the emergence of a pathogen recognition repertoire in a way comparable to the adaptive immune systems of vertebrates. The mammal antigen receptor repertoire is generated via the Variable-Diversity-Joining (VDJ) rearrangement of immunoglobulin segments, which is mediated by the RAG (recombination activating gene) transposon. In jawless fishes, somatic rearrangement of LRR genomic cassettes generates the variable lymphocyte receptors, which are structurally extremely similar to the LRR domain of the Ectocarpus ROCO proteins. In Ectocarpus, intronic and other LRRs scattered in the genome could similarly serve as a source of diversity for the LRR exon shuffling. The Ectocarpus ROCO and NB-ARC families also share some genomic features with plant resistance genes, in particular, physical clustering and high gene birth and death rates. Patterned rectangles and circles represent hypervariable ligand-binding LRR and TPR modules (respectively) subject to positive selection. Curved arrows represent exon shuffling events followed by divergence. Light gray rectangles highlight domains likely involved in signal transduction events triggered by recognition.