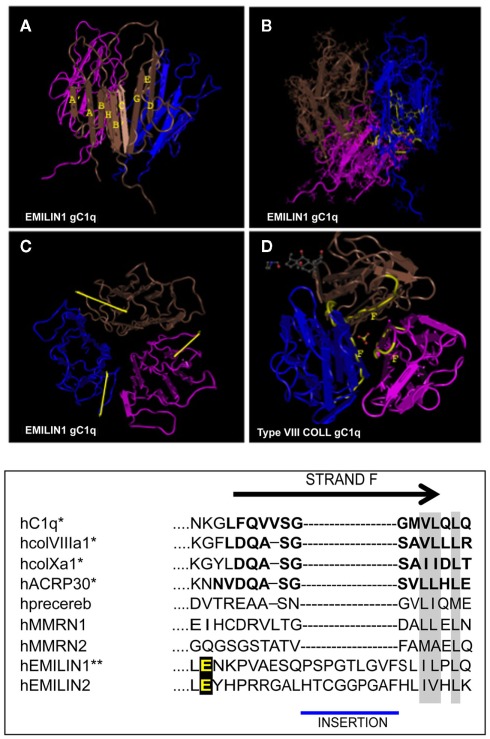
Figure 2.
Top: NMR solution structure of the homotrimeric EMILIN1 gC1q domain. The structure was downloaded from database of protein structures maintained at NCBI site (http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid = 68072). Ribbon representation of the assembly, as side view, is presented in (A) the three protomers in the trimer are shown in different colors (pink, blue, and brown). Each monomer has a nine-stranded folding topology, with strands labeled according to the gC1q/tumor necrosis factor superfamily nomenclature. (B) The residues highly conserved in all the members of the gC1q superfamily and essential for a correct domain folding are shown in yellow only in one monomer for clarity. (C) Top view of EMILIN1 gC1q domain. The yellow bar highlights the solvent exposed position of the unstructured segment Tyr927–Gly945 (D) the X-ray structure of type VIII collagen gC1q, http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid = 25284, showing the buried position of strand F. The same position of strand F is present in the X-ray solution structures of ACRP, type X collagen and Complement gC1q domains. Bottom: Sequence alignment between strand F (residues in bold) of representative members of the C1q superfamily and EMILINs gC1q. Asterisks indicate proteins for which the structure has been solved. Similar residues that are conserved in all proteins are shaded in gray, and the glutamic acid residue interacting whit α4β1 is in yellow.