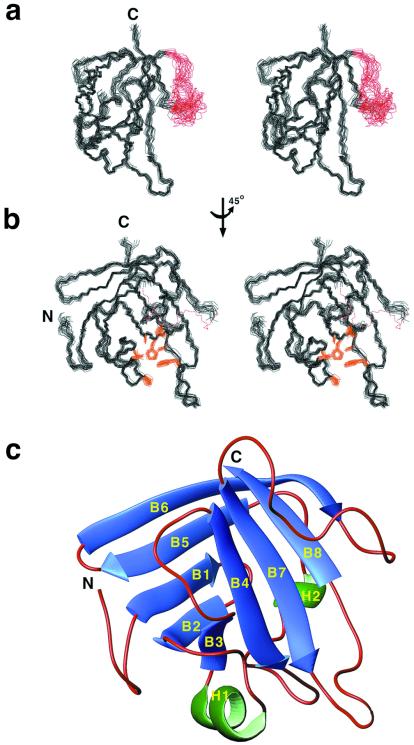
Figure 3.
SrtAΔN59 adopts a unique three-dimensional β-barrel structure. (a) Wall-eyed stereoview of the ensemble of 25 SrtAΔN59 conformers. Residues K62 to V161 and K175 to V205 (amino acid numbering is for full-length sortase) are well ordered in solution (black). The structurally disordered loop (K162–G174) connecting strands β5 and β6 is colored red. (b) Amino acids within the active site are well defined by the NMR data. The panel shows the ensemble of conformers rotated by 45° degrees with the active-site side chains colored gold (A92, P94, L97, A118, H120, I182, C184, W194). The β5–β6 loop is represented by a red dashed line. (c) Ribbon drawing of the structure of SrtAΔN59. β-strands and helices are colored blue and green, respectively. Beginning at the N terminus, the β1 strand (G74–I78) is followed by a short hairpin and lies antiparallel to strand β2 (I83–Y88). The β2 and β3 strands (V101–A104) are positioned in parallel and are tethered by a 310 helix (P94–L97) which crosses over the surface of the enzyme to form the lateral wall of the active site. Strands β3 and β4 (Q113–G119) lie antiparallel with respect to one another and are followed by a long loop and a second α-helix, which assume a circuitous path to position strand β5 (S140–V146) for antiparallel alignment with strand β1. Strand β6 (E149–K155) is then connected by a short hairpin turn for antiparallel pairing, followed by a long loop structure to connect it to strand β7 (K177–T183), which is aligned parallel to β4. The active-site sulfhydryl, C184, is positioned at the end of β7, which also includes LITC184, the signature sequence of sortase enzymes. The structure is completed by a loop (D185–W194), which connects strands β7 and β8 (E195–F200) for antiparallel pairing. Analysis with the program dali reveals no structural homologs (no structures with Z scores >2.1).