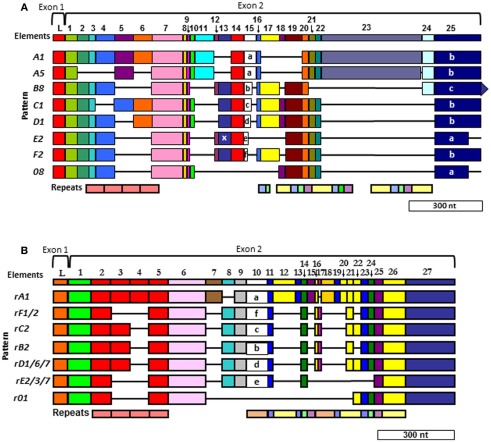
Figure 6.
Two different alignments for Sp185/333 sequences are equally optimal. (A) The cDNA alignment was initially done with ESTs and full-length cDNA sequences (Terwilliger et al., 2006, 2007). (B) The Repeat-based alignment optimizes correspondence between repeats and elements whenever possible (Buckley and Smith, 2007). Optimal alignments require the insertion of artificial gaps (horizontal black lines) that delineate individual elements shown as colored blocks (the consensus of all elements are numbered across the top of each alignment; L, leader). Different element patterns are based on the variable presence or absence of elements. Designations of element patterns are listed to the left of each alignment. There are three types of element 25 in (A) (a, b, and c), which are defined by the location of three possible stop codons. A common, single nucleotide RNA edit in element pattern E2 alters a glycine codon to a stop (X) in element 13 and is denoted as an E2.1 pattern. Repeats, shown at the bottom of (A,B), are shows as different colors and occur as tandem repeats and interspersed tandem repeats. The genes have two exons (brackets at the top of each alignment), of which the first encodes the leader and the second encodes the mature protein including all of the elements. A single intron of ∼400 nt is positioned between the leader and the first element and is not shown. Modified from Ghosh et al. (2010) with permission from Elsevier.