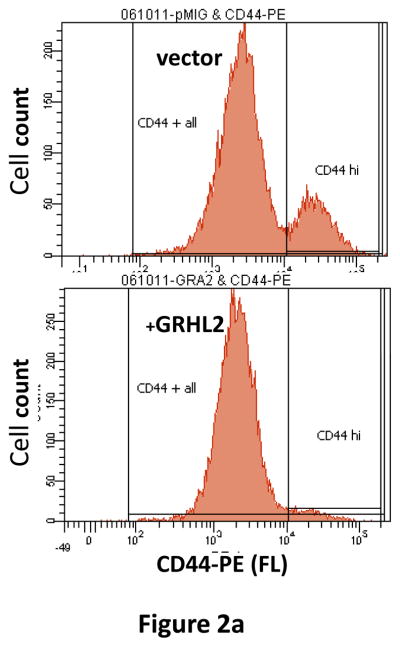
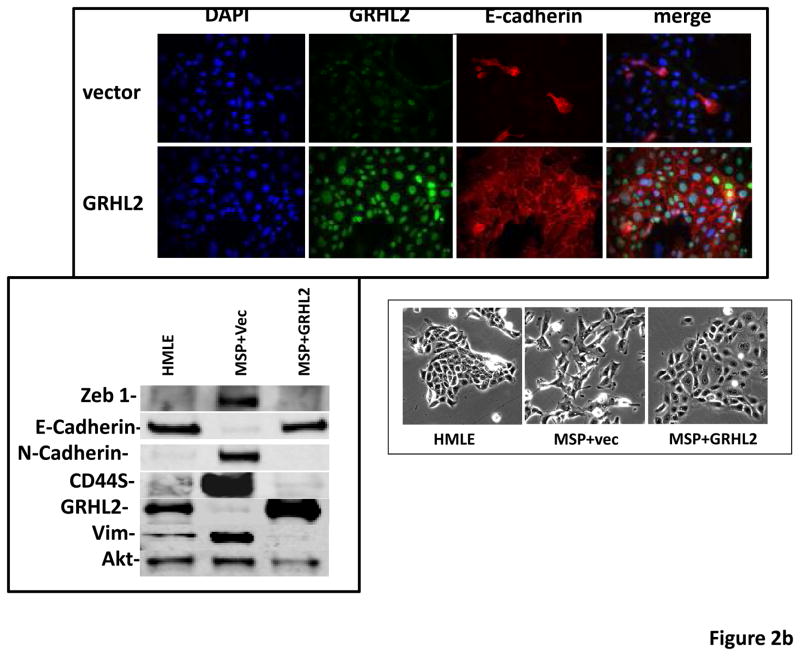
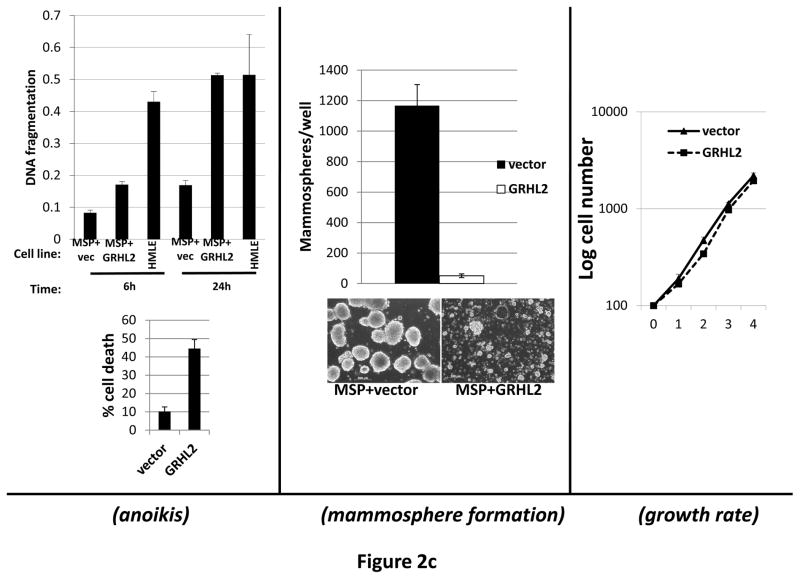
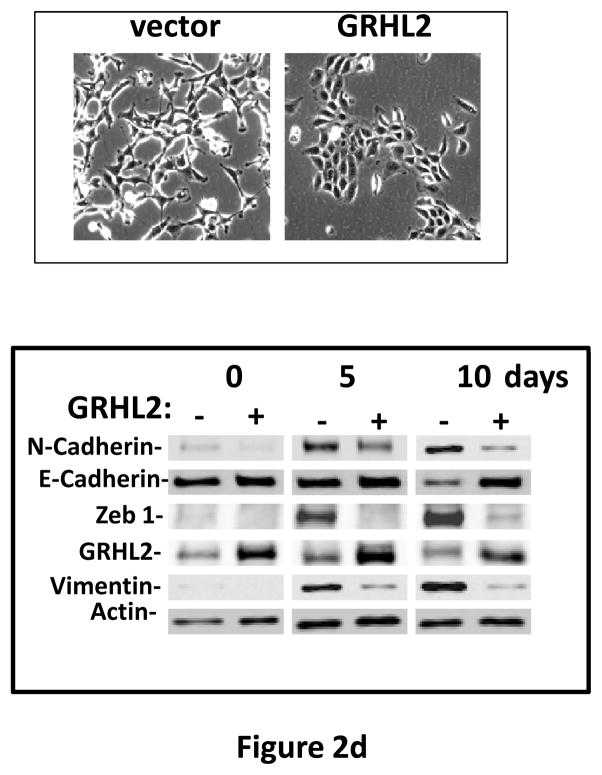
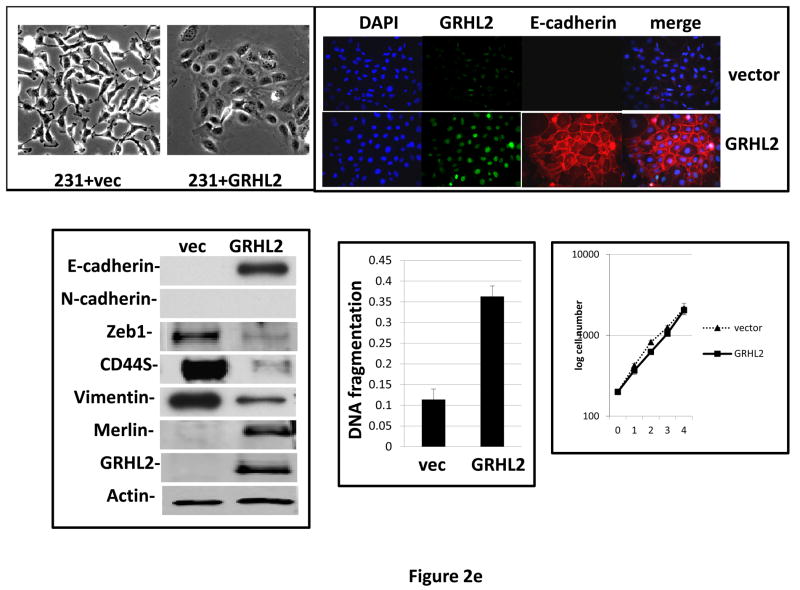
Figure 2. GRHL2 suppresses EMT.
(a) Ectopic expression of GRHL2 suppresses the CD44 expression in the spontaneously occurring mesenchymal subpopulation of HMLE cells (“MSP”). HMLE+Twist-ER cells (without 4-OHT) infected with GRHL2 or empty retroviral vector (pMIG) were analyzed for CD44 expression by FACS. (b) GRHL2 reverts MSP cells to an epithelial phenotype. The CD44high mesenchymal sub-population of HMLE, obtained by flow-sorting of HMLE cells, were infected with empty vector or GRHL2 and then (top panel) stained for the indicated proteins by immunofluorescence or (lower panel) probed for epithelial and mesenchymal marker genes by western blotting. (c) GRHL2 expression in MSP cells restores anoikis-sensitivity (left panels: top graph represents a DNA fragmentation ELISA assay, lower graph represents a trypan blue-permeability assay) and reduces mammosphere (middle panels), without affecting growth rate (right panel). (d) GRHL2 suppresses Twist-induced EMT. HMLE+twist-ER expressing either empty vector or GRHL2 were treated with 4-OHT (to activate the twist transgene) for 7 days, and (top panel) photographed to record morphology. (lower panel): Time course of changes in epithelial and mesenchymal marker genes after induction of the twist gene with 4-OHT, in cells with or without ectopic GRHL2 expression; (e) GRHL2 suppresses EMT and anoikis-resistance in MDA-MB-231LN cells. MDA-MB-231LN expressing either empty vector or GRHL2 were analyzed for morphology (phase contrast microscopy, top left), E-cadherin expression and localization (immunofluorescence, top right), expression of epithelial and mesenchymal markers (western blotting, bottom left), anoikis-sensitivity (DNA fragmentation ELISA (middle) and adherent growth rate (lower right).