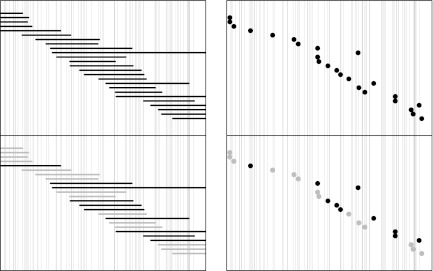
FIG. 6.
Schematic representation of “thinning” in Rmap alignments. The horizontal axis represents the underlying genome, with vertical lines indicating restriction sites. (Left) Rmaps are represented as intervals. (Right) They are viewed as point events represented by the midpoint of the Rmaps. (Top) The top panel in both plots represent the true shotgun random sample of Rmaps that originated from this region. Actual Rmaps obtained by image processing will have noise, including sizing errors, missing cuts and false cuts, so not all these Rmaps will be successfully aligned. Further, the chance of being aligned may depend on the location of the Rmap; for example, Rmaps with fewer fragments (from regions with fewer recognition sites) may be less likely to align than Rmaps of similar length with more fragments. (Bottom) In the bottom panels, which represent the results of alignment, unaligned Rmaps are indicated in gray. Since the probability of being successfully aligned depends on the origin, the locations of aligned Rmaps, which is what we actually observe, are no longer uniformly distributed.