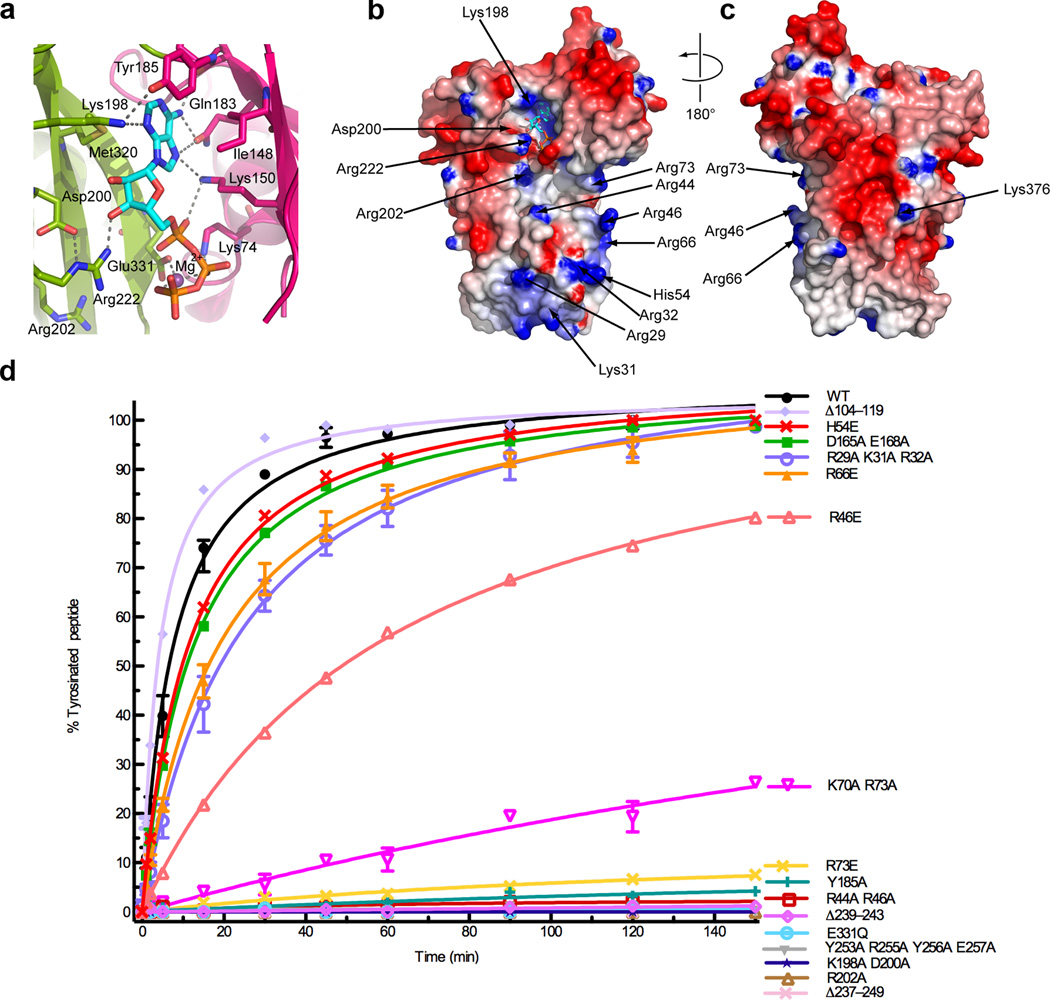
Figure 2.
Active site architecture and α–tubulin C-terminal peptide recognition. (a) Conserved interactions in the TTL active site, colored as in Fig. 1a (b) Nucleotide (ventral) and (c) dorsal views of the TTL molecular surface color-coded for electrostatic potential55 (red, negative; blue, positive, ranging from −7 kBT to 7 kBT) (d) Tyrosination of a 14-residue α–tubulin C-terminal peptide by TTL and structure-guided TTL mutants (N≥2; Supplementary Fig. 3). The 14-residue peptide (VDSVEGEGEEEGEE) serves as an optimal TTL peptide substrate42. Error bars indicate s.e.m. and are frequently smaller than the symbols.