Abstract
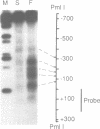
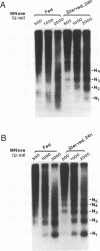
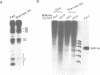
Transcription of the chicken malic enzyme gene in the liver is stimulated by feeding and inhibited by starvation. Concomitant with the increase in transcription caused by refeeding, chromatin structure around the transcription start site of the malic enzyme gene is modified in the liver. Digestion of chromatin in isolated nuclei with DNase I revealed four feeding-induced DNase I hypersensitive sites (-220, -170, -130 and -70 bp) near the malic enzyme promoter. Similarly, digestion of chromatin with restriction endonucleases detected enhanced cleavage within this region when birds were refed. Micrococcal nuclease detected the presence of nucleosomes over this region in the starved state, but not in the fed state. After food was withdrawn from fed birds, nucleosomes were reformed in this region within 6 h. The speed and magnitude of the changes in nucleosomal structure in this region suggest that they did not require DNA replication.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almer A., Rudolph H., Hinnen A., Hörz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986 Oct;5(10):2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbarić S., Fascher K. D., Hörz W. Activation of the weakly regulated PHO8 promoter in S. cerevisiae: chromatin transition and binding sites for the positive regulatory protein PHO4. Nucleic Acids Res. 1992 Mar 11;20(5):1031–1038. doi: 10.1093/nar/20.5.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker P., Renkawitz R., Schütz G. Tissue-specific DNaseI hypersensitive sites in the 5'-flanking sequences of the tryptophan oxygenase and the tyrosine aminotransferase genes. EMBO J. 1984 Sep;3(9):2015–2020. doi: 10.1002/j.1460-2075.1984.tb02084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellard M., Dretzen G., Giangrande A., Ramain P. Nuclease digestion of transcriptionally active chromatin. Methods Enzymol. 1989;170:317–346. doi: 10.1016/0076-6879(89)70054-9. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Bresnick E. H., Bustin M., Marsaud V., Richard-Foy H., Hager G. L. The transcriptionally-active MMTV promoter is depleted of histone H1. Nucleic Acids Res. 1992 Jan 25;20(2):273–278. doi: 10.1093/nar/20.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresnick E. H., John S., Hager G. L. Histone hyperacetylation does not alter the positioning or stability of phased nucleosomes on the mouse mammary tumor virus long terminal repeat. Biochemistry. 1991 Apr 9;30(14):3490–3497. doi: 10.1021/bi00228a020. [DOI] [PubMed] [Google Scholar]
- Cordingley M. G., Riegel A. T., Hager G. L. Steroid-dependent interaction of transcription factors with the inducible promoter of mouse mammary tumor virus in vivo. Cell. 1987 Jan 30;48(2):261–270. doi: 10.1016/0092-8674(87)90429-6. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992 Jan 16;355(6357):219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- Goodridge A. G. Dietary regulation of gene expression: enzymes involved in carbohydrate and lipid metabolism. Annu Rev Nutr. 1987;7:157–185. doi: 10.1146/annurev.nu.07.070187.001105. [DOI] [PubMed] [Google Scholar]
- Grange T., Roux J., Rigaud G., Pictet R. Two remote glucocorticoid responsive units interact cooperatively to promote glucocorticoid induction of rat tyrosine aminotransferase gene expression. Nucleic Acids Res. 1989 Nov 11;17(21):8695–8709. doi: 10.1093/nar/17.21.8695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. Nucleosomes: regulators of transcription. Trends Genet. 1990 Dec;6(12):395–400. doi: 10.1016/0168-9525(90)90299-l. [DOI] [PubMed] [Google Scholar]
- Jump D. B., Bell A., Santiago V. Thyroid hormone and dietary carbohydrate interact to regulate rat liver S14 gene transcription and chromatin structure. J Biol Chem. 1990 Feb 25;265(6):3474–3478. [PubMed] [Google Scholar]
- Kornberg R. D., Lorch Y. Irresistible force meets immovable object: transcription and the nucleosome. Cell. 1991 Nov 29;67(5):833–836. doi: 10.1016/0092-8674(91)90354-2. [DOI] [PubMed] [Google Scholar]
- Ma X. J., Salati L. M., Ash S. E., Mitchell D. A., Klautky S. A., Fantozzi D. A., Goodridge A. G. Nutritional regulation and tissue-specific expression of the malic enzyme gene in the chicken. Transcriptional control and chromatin structure. J Biol Chem. 1990 Oct 25;265(30):18435–18441. [PubMed] [Google Scholar]
- Reik A., Schütz G., Stewart A. F. Glucocorticoids are required for establishment and maintenance of an alteration in chromatin structure: induction leads to a reversible disruption of nucleosomes over an enhancer. EMBO J. 1991 Sep;10(9):2569–2576. doi: 10.1002/j.1460-2075.1991.tb07797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaud G., Roux J., Pictet R., Grange T. In vivo footprinting of rat TAT gene: dynamic interplay between the glucocorticoid receptor and a liver-specific factor. Cell. 1991 Nov 29;67(5):977–986. doi: 10.1016/0092-8674(91)90370-e. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Nucleosome positioning: occurrence, mechanisms, and functional consequences. Prog Nucleic Acid Res Mol Biol. 1991;40:143–184. doi: 10.1016/s0079-6603(08)60841-7. [DOI] [PubMed] [Google Scholar]
- Svaren J., Chalkley R. The structure and assembly of active chromatin. Trends Genet. 1990 Feb;6(2):52–56. doi: 10.1016/0168-9525(90)90074-g. [DOI] [PubMed] [Google Scholar]
- Travers A. A. DNA conformation and protein binding. Annu Rev Biochem. 1989;58:427–452. doi: 10.1146/annurev.bi.58.070189.002235. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Yamamoto K. R. Reversible and persistent changes in chromatin structure accompany activation of a glucocorticoid-dependent enhancer element. Cell. 1984 Aug;38(1):29–38. doi: 10.1016/0092-8674(84)90523-3. [DOI] [PubMed] [Google Scholar]