Figure 5. IGCR1 mediates long distance IgH chromosomal loops.
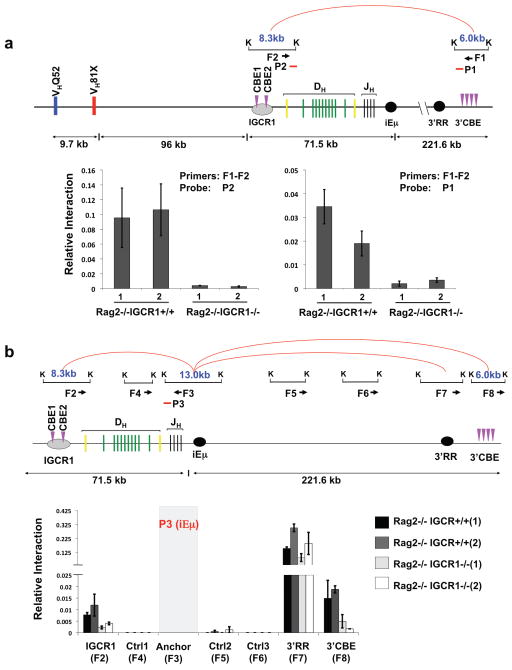
(a) Schematic of chromosome interactions between IGCR1-containing and 3′IgHCBE-containing KpnI restriction fragments in 3C assays. Interactions between IGCR1 and 3′IgHCBE locales in 129SV RAG2-/- and RAG2-/-IGCR1-/- A-MuLV transformed pro-B cells were quantified by real time PCR (Taqman) using probe (P2) (left) and a probe (P1) (right). (b) Schematic of chromosome interactions between iEμ-containing KpnI restriction fragment and indicated KpnI restriction fragments in other IgH locales. Interactions between iEμ and IGCR1, iEμ and 3′IgHRR locales, iEμ and 3′IgHCBE locales in RAG2-/- and RAG2-/-IGCR1-/- A-MuLV transformed pro-B cells were quantified by real time PCR using a probe (P3) from the iEμ locale. F1-F8 indicate primers used for PCR. K indicates KpnI sites. Red arcs indicate interactions detected in RAG2-/- cells. The average association frequency of three independent 3C experiments with 2 independent A-MuLV transformed lines from each genotype is shown with standard deviation indicated.
