Fig. 9.—
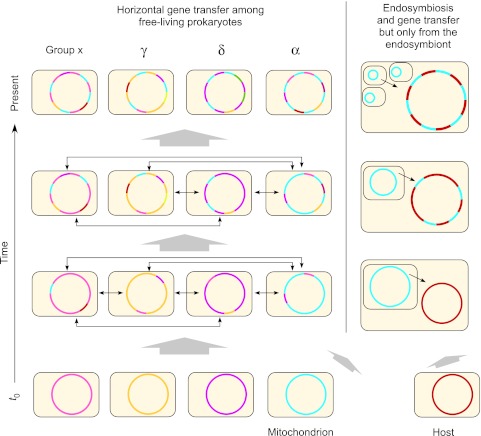
Lateral gene transfer between free-living prokaryotes subsequent to the origin of organelles requires that we think at least twice when interpreting phylogenetic trees for genes that were acquired from mitochondria (or chloroplasts, not shown). Genes that entered the eukaryotic lineage via the genome of the mitochondrial endosymbiont represent a genome-sized sample of prokaryotic gene diversity that existed at the time that mitochondria arose. The uniformly colored chromosomes at t0 indicate that at the time of mitochondrial origin, there existed for individual prokaryotes specific collections of genes in genomes, much like we see for strains of Escherichia coli today. If an E. coli cell would become an endosymbiont today, it would not introduce an E. coli pangenome’s worth of gene diversity (some 18,000 genes) into its host lineage, rather it would introduce some 4,500 genes or so. The free-living relatives of that endosymbiont would go on reassorting genes across chromosomes via gene transfer at the pangenome (species and strain) level, at the genus level, at the family level, and at the level of proteobacteria, the environment, and so forth. After 1.5 billion years, it would be very unreasonable to expect any contemporary prokaryote to harbor exactly the same collection of genes as the original endosymbiont did. Instead, the descendant genes of the endosymbiont (labeled blue in the figure) would be dispersed about myriad chromosomes, and we would eventually find them one at a time through genome sequencing of individuals from different groups. Though not shown here, for reasons of space limitation, exactly the same process also applies, in principle, for the host’s genome. Redrawn from Martin (1999b) and from figure 5 of Rujan and Martin (2001).