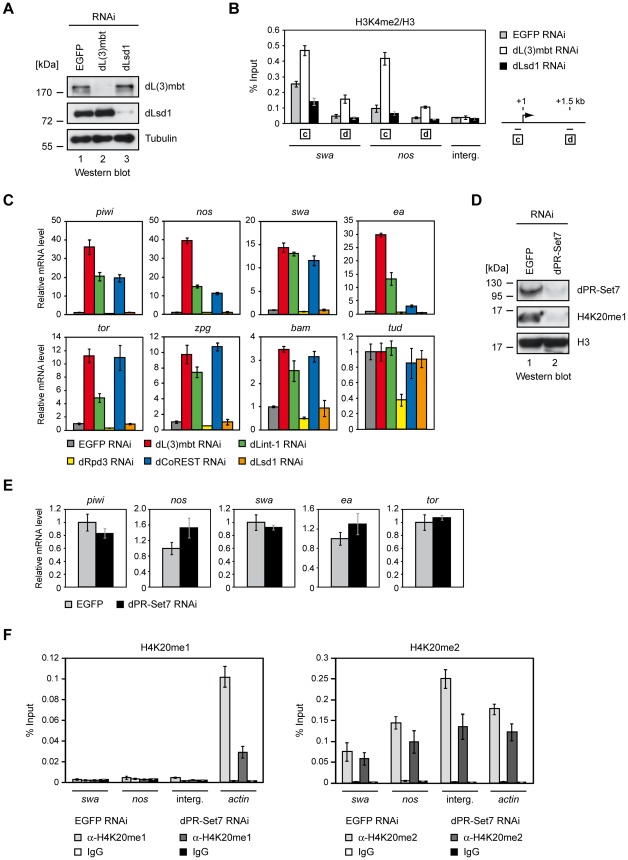
Figure 6. dLsd1, dRpd3, and dPR-Set7 are not essential for MBTS gene repression.
(A) Kc cells were treated with dsRNA directed against EGFP, dL(3)mbt and dLsd1. Nuclear extracts of RNAi treated Kc cells were subjected to Western blot and analyzed using antibodies as indicated. (B) Chromatin of RNAi treated cells was precipitated with H3K4me2 and H3 antibodies as indicated. The ratio of H3K4me2 and H3 ChIP signals is shown for swa and nos promoter and ORF regions and an unrelated intergenic region (interg.). Genes analyzed are denoted below the panel. Amplified regions are indicated by boxed letters and have the following distances from the transcriptional start site as illustrated on the right: c, 0–0.15 kb (promoter); d, 1.5 kb downstream. (C) Kc cells were treated with dsRNA directed against EGFP, dL(3)mbt, dLint-1, dRpd3, dCoREST and dLsd1 as indicated and transcription was determined by RT-qPCR. Transcription levels in EGFP RNAi treated cells were set to 1. Tudor serves as a negative control. (D) Kc cells were treated with dsRNA directed against EGFP and dPR-Set7 as indicated. Nuclear extracts and acid extracted histones were analyzed by Western blot as indicated. (E) Transcription levels of target genes were determined by RT-qPCR. Transcription levels in cells treated with dsRNA against EGFP were set to 1. (F) Chromatin from cells treated with RNAi against dPR-Set7 or EGFP (control) was precipitated with H4K20me1 and IgG (left panel) or H4K20me2 and IgG (right panel) antibodies as indicated. ChIP signals are shown for swa and nos promoter regions, an intergenic region and the actin gene as denoted below the panel.