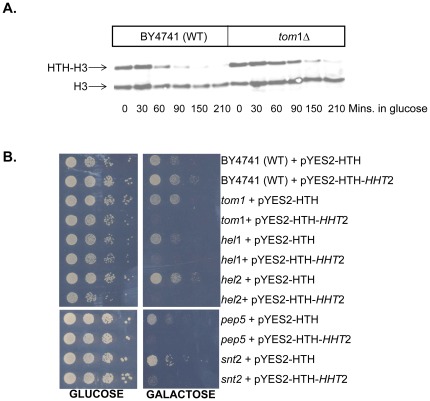
Figure 1. Identification of strains lacking specific E3 ligases that confer sensitivity to histone overexpression.
(A) The tom1 deletion strain can degrade exogenous histones with much slower kinetics than the wild type strain. Histone degradation assay was carried out exactly as described previously [7] whereby a galactose inducible, HIS10-TEV-HA (HTH) tagged histone H3 was expressed in G1 arrested cells for 90 minutes, following which cells were switched to glucose media and harvested at the indicated time points. Whole cell extracts were prepared and resolved on 18% polyacrylamide gels and subjected to Western blotting with the H3-C antibody described previously [6] that recognizes both the endogenous chromatin associated histone H3 as well as the overexpressed H3. The entire experiment was carried out in G1 arrested cells to prevent incorporation of exogenous histones into chromatin that occurs in replicating cells. (B.) Strains lacking Hel1, Hel2, Pep5 and Snt2 E3 ligases are sensitive to histone overexpression. The histone overexpression sensitivity screen was carried out exactly as described previously [7]. Briefly, wild type (WT) BY4741 and deletion strains corresponding to the indicated E3 ligases were transformed with either an empty vector (pYES2-HTH) or a galactose inducible HTH tagged histone H3 expressing plasmid (pYES2-HTT-HHT2). Then 10-fold serial dilutions of the resulting transformants were plated on glucose (no histone overexpression) or galactose (allows histone overexpression) containing media without uracil to select for the plasmid. Plates were photographed after 3 days of incubation at 30°C.