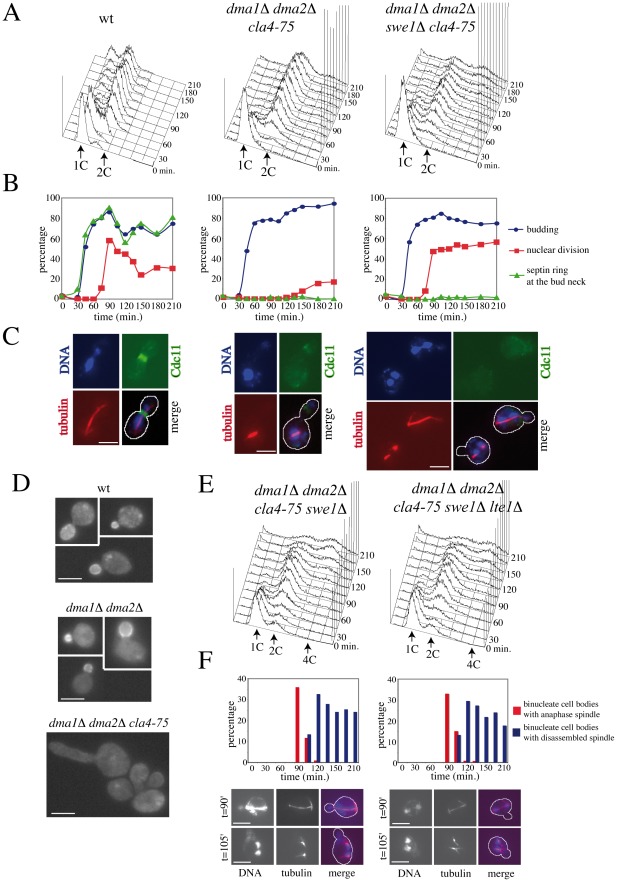
Figure 4. Morphogenesis checkpoint and SPOC response to spindle mispositioning in cells lacking Dma proteins and Cla4.
A–C: Logarithmically growing cultures of wild type (W303), dma1Δ dma2Δ cla4-75 (ySP5247) and dma1Δ dma2Δ cla4-75 swe1Δ (ySP6238) cells were arrested in G1 by alpha factor at 25°C and released into the cell cycle at 37°C at time 0. Cells were collected at the indicated times for FACS analysis of DNA contents (A) and scoring of budding, nuclear division, spindle assembly and septin ring assembly by immunofluorescence with anti-tubulin and anti-Cdc11 antibodies, respectively (B). Representative micrographs were taken 90 minutes after release (C). Scale bars: 5 µm. D: Localization of Lte1-GFP in wild type (ySP3333), dma1Δ dma2Δ (ySP4380) and dma1Δ dma2Δ cla4-75 (ySP7766) cells expressing Lte1-GFP from the GAL1 promoter after induction with 1% galactose for 1 hour followed by shift to 37°C for 3 hours. Scale bars: 5 µm. E–F: Cell cultures of dma1Δ dma2Δ cla4-75 swe1Δ (ySP6238) and dma1Δ dma2Δ cla4-75 swe1Δ lte1Δ (ySP7672) cells were treated as above. E: FACS analysis of DNA contents. F: the percentage of cells undergoing mispositioned anaphase (i.e. binucleate cell bodies with or without an intact anaphase spindle) was calculated over the total amount of binucleate cells. Nuclei were stained with DAPI and spindles were visualized by immunostaining of tubulin. Micrographs show representative cells undergoing anaphase in the mother cell and with an intact (t = 90′) or a disassembled spindle (t = 105′). Scale bars: 5 µm.