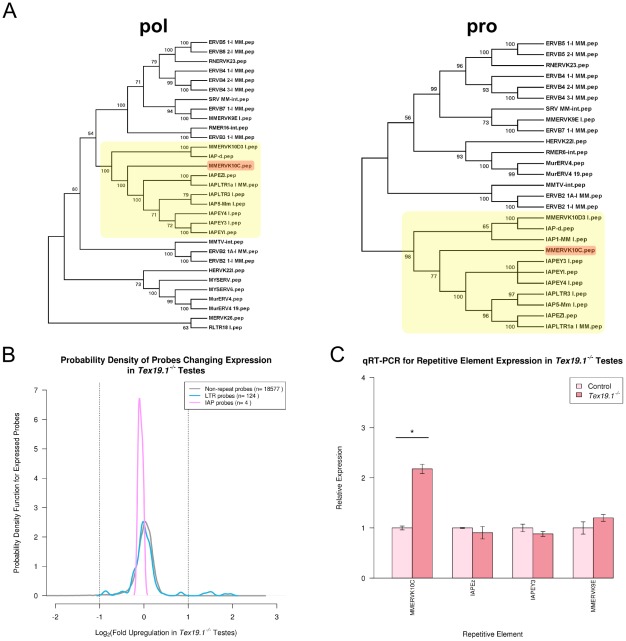
Figure 2. Closely related retrotransposons are differentially sensitive to loss of Tex19.1.
(A) Phylogeny of mouse retrotransposon pol and pro proteins. MMERVK10C sequences are highlighted in red. The MMERVK10C sequences lie within a cluster of IAP-type sequences (yellow). (B) Plot showing the likelihood of IAP probes changing expression in the Tex19.1−/− microarray dataset. (C) qRT-PCR for retrotransposons closely related to MMERVK10C in Tex19.1−/− knockout and littermate control testes at 16 dpp. Expression levels for each repetitive element (mean ± standard error for three animals) were normalized to β-Actin and expressed relative to littermate controls. MMERVK10C env.c and IAP primer sets (Figure S2) were used to assess MMERVK10C and IAPEz expression. Asterisk indicates a statistically significant difference (p<0.05).