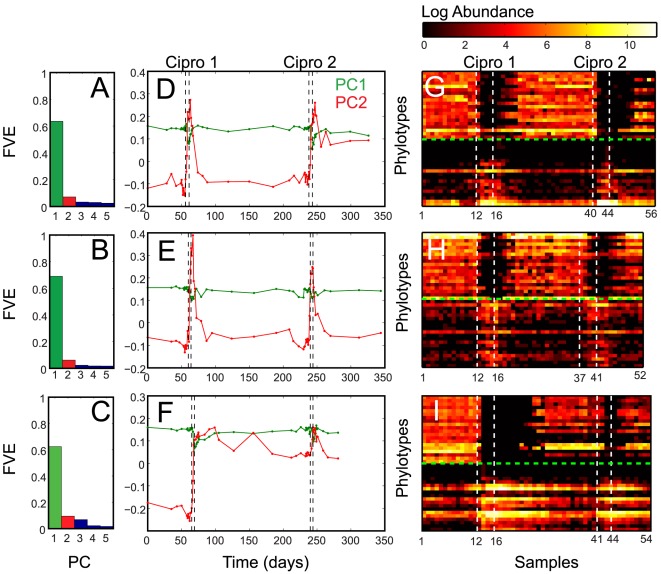
Figure 5. Analysis of microbiota response to the antibiotic ciprofloxacin from three subjects [24] using singular value decomposition identifies antibiotic-sensitive and antibiotic-tolerant bacteria.
A–C: fraction of variance explained by the five most dominant components. D–F: plot of each sample component 1 (green) and 2 (red) coordinates versus sample time. G–I: sorting of the phylotypes log2-transformed abundance matrix based on the correlation within the two principal component. Above (below) the green dashed lines, we display the time series of the top 20 phylotypes strongly correlated (anti-correlated) with component 1 and anti-correlated (correlated) with 2 and dropping (increasing) during treatment, which we identify as sensitves (tolerants). Subject 3 (C,F,I) displays absence of sensitive bacteria for a prolonged period of about 50 days after the first antibiotic treatment. This confirms the fact that microbiota response to antibiotic can differ from subject to subject. Additionally, it also supports our model prediction of remaining locked in a tolerant-dominated state after antibiotic treatment cessation.