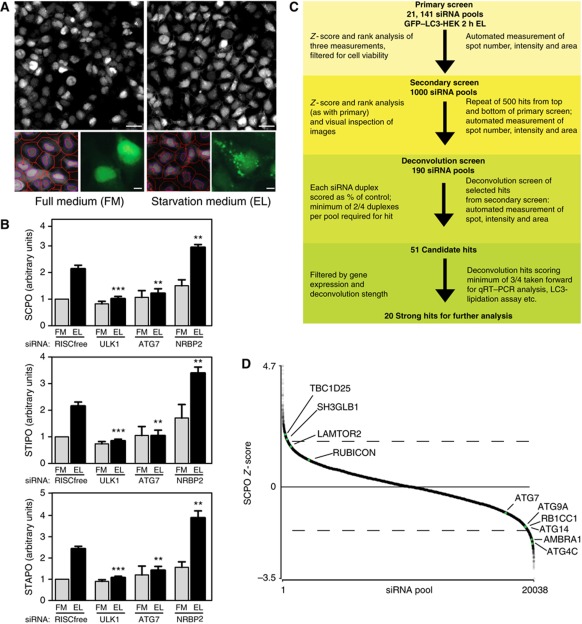
Figure 1.
Genome-wide screen for starvation-induced autophagy. (A) Cellomics images (top) of GFP–LC3-HEK cells in full medium (FM) or after 2 h in starvation medium (ES) with leupeptin (EL). Scale bars=40 μm. Optimised automated analysis (bottom left) programme detecting hoechst-labelled nuclei (purple line), cell outline (red line) and identifies GFP–LC3 spots (red spots) in FM and EL. Enlarged GFP–LC3 images in FM and EL (bottom right). Scale bars=10 μm. (B) SCPO, STIPO and STAPO after indicated siRNA treatment of GFP–LC3-HEK cells incubated as in (A). Error bars represent s.e.m. Significance was determined using a two-tailed paired t-test compared with RISCfree (RF) in EL: SCPO siULK1, ***P=0.0002; siATG7, **P=0.0078; siNRBP2, **P=0.0088. STIPO siULK1, ***P<0.0001; siATG7, **P=0.0060; siNRBP2, **P=0.0041. STAPO siULK1, ***P<0.0001; siATG7, **P=0.0037; siNRBP2, **P=0.0028. The experiments were performed: RISCfree (n=5), siULK1 (n=5), siATG7 (n=3), siNRBP2 (n=3). (C) Overview of screening strategy. (D) Scatter plot of median SCPO Z-scores from siRNA pools in the primary screen that met the minimum object count (OC) cutoff. Dashed lines represent approximate cutoff for 500 best increasers and decreasers. RP of known autophagy genes (ATG4C, ATG14, RB1CC1, ATG9A, ATG7) are given in Supplementary Table S2; AMBRA1 (FLJ20294), TBC1D25 (OATL1) and SH3GLB1 (BIF-1) in Supplementary Table S1; LAMTOR2 has a RP=708.7 and RUBICON has a RP=1126.