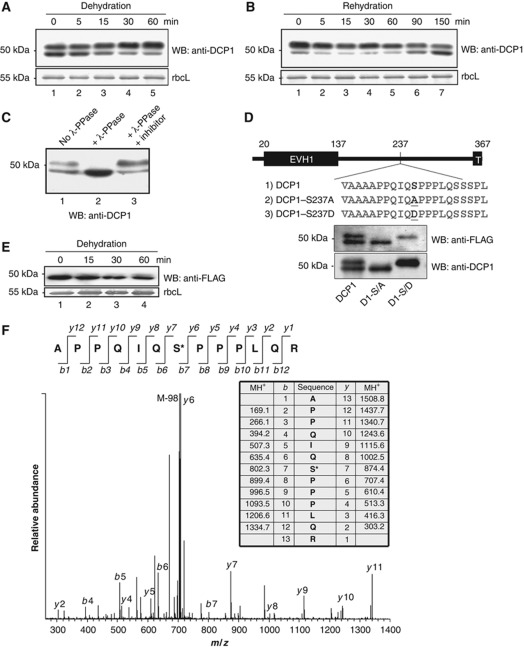
Figure 1.
Characterization of DCP1 phosphorylation site. (A) Western blot analysis of DCP1 from seedlings subjected to dehydration stress. Samples were taken at different times after dehydration as indicated. Coomassie blue-stained rbcL (large subunit of ribulose-1,5-bisphosphate carboxylase) was used as a loading control. (B) DCP1 from dehydrated seedlings subjected to rehydration. (C) Western blot analysis of DCP1. Samples were either untreated or treated with λ phosphatase (λ-PPase) or with λ-PPase plus inhibitor mix. (D) Schematic presentation and western blot analysis of DCP1 and its S237 mutants in complementation lines. The phosphorylation site and mutated sites were highlighted. EVH1, Enabled/VASP homology 1. T, Trimerization domain. (E) Western blot analysis of transgenic plants expressing FLAG–DCP1-S237A subjected to dehydration stress. (F) MS–MS spectrum of the phosphorylated DCP1 peptide containing the phosphorylation site at S237. Phosphorylated DCP1 was digested with trypsin followed by LC-MS–MS analysis. The monoisotopic mass of the phosphopeptide ion was calculated at 1508.8 (MH1+). The fragment ion at 705.89 (MH22+) displays a strong ion signal intensity corresponding to the loss of phosphoric acid (98) from the precursor ion. Top panel shows the b- and y-fragment ions of the peptide. Both b- series and y-ion series confirmed S237 to be the phosphorylation site.