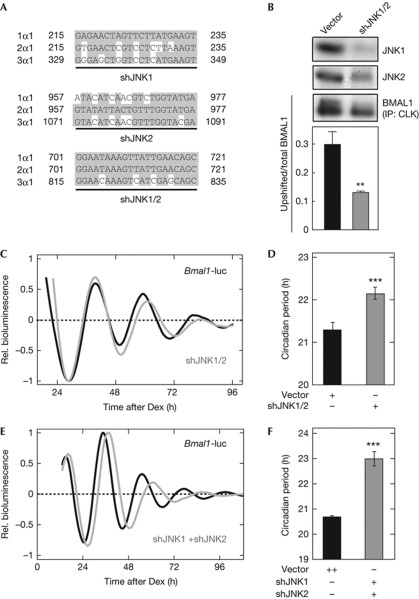
Figure 3.
Effects of knockdown of c-Jun N-terminal kinase (JNK)1 and JNK2 on BMAL1 phosphorylation and cellular rhythms. (A) Two shRNA constructs, shJNK1 and shJNK2, respectively, target JNK1 and JNK2, while shJNK1/2 targets both JNK1 and JNK2. Nucleotide numbers starting from initiation codon are shown. (B) NIH3T3 cells were transfected with shJNK1/2 or the empty vector. After 48-h incubation, the cells were synchronized by 2-h dexamethasone (Dex) pulse. The cells were collected 24 h after the synchronization, and were subjected to immunoprecipitation (IP) and immunoblot analysis. Data are means with s.e.m. (n=4; **P<0.01, Student's t-test, versus empty vector). (C) Cellular rhythms were recorded from the NIH3T3 cells transfected by 0.5 μg of Bmal1us0.3/pGL4 with 0.5 μg of shJNK1/2 (grey line) or the empty vector pBS-mU6 (black line). (D) The circadian periods with and without shJNK1/2 were 22.15±0.14 and 21.31±0.16 h, respectively. Data are means with s.e.m. (n=10; ***P<0.001, Student's t-test, versus empty vector). (E) NIH3T3 cells were transfected by 0.5 μg of Bmal1us0.3/pGL4 in combination with 0.5 μg each of shJNK1/pBS-mU6 and shJNK2/pBS-mU6 (grey line) or 1 μg of the empty vector pBS-mU6 (black line). Cellular rhythms were synchronized by 2-h Dex treatment, and the time point of the medium change to the recording medium was defined as time 0. Shown were the detrended curves of bioluminescence rhythms. (F) Circadian periods of the rhythms with and without shJNK1 and shJNK2 were 22.99±0.28 and 20.68±0.05 h, respectively. Data are means with s.e.m. (n=4; ***P<0.001, Student's t-test, versus empty vector).