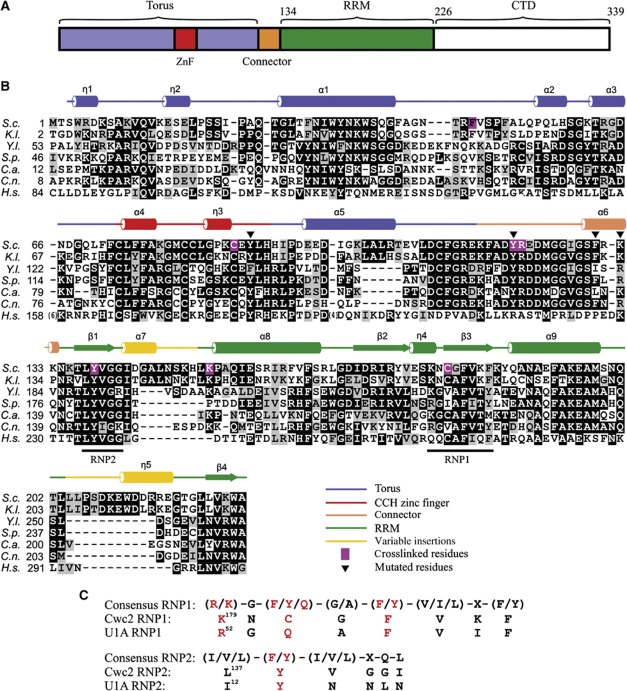
Figure 3.
Domain composition and sequence alignment of Cwc2. (A) Schematic depiction of domains and structural motifs in Cwc2. (B) Multiple sequence alignment of Cwc2 orthologues. S.c.—S. cerevisiae; K.l.—Kluyveromyces lactis; Y.l.—Yarrowia lipolytica; S.p.—Schizosaccharomyces pombe; C.a.—Candida albicans; C.n.—Cryptococcus neoformans; H.s.—Homo sapiens. Darker background corresponds to higher sequence conservation. Icons above indicate secondary structure elements from the Cwc2 crystal structure. Magenta background indicates the RNA-crosslinked sites. The triangles indicate mutated amino acids tested in splicing complementation assays. (C) Comparison of consensus RNP1 and RNP2 sequences (Maris et al, 2005) in Cwc2 and U1A, respectively. The amino-acid residues important for RNA binding are depicted in red. X represents any amino acid.