Figure 6. Sequence dependent ASM reveals sequence determinants of DNA methylation.
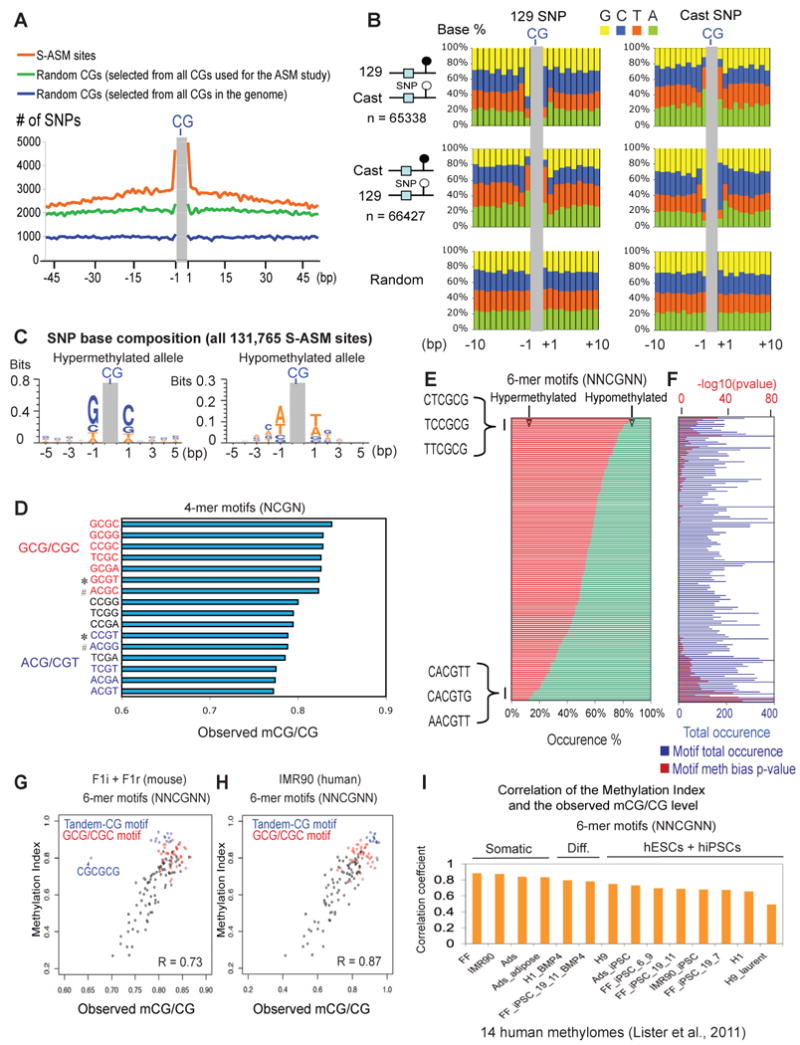
(A) The total number of SNPs at each base within +/- 50bp of 131,765 sequence dependent ASM sites (orange) is shown. A similar plot is shown for 131,765 random CG sites drawn from either all CGs selected for the ASM study (green) or all CGs in the whole genome (blue). (B) The SNP base composition on the 129 (left) or the Cast (right) allele is shown for those near ASM sites that are preferentially methylated on the 129 allele (top) or the Cast allele (middle). A similar analysis was done for a control set of random CGs of equal size drawn from all CGs selected for the ASM study (bottom). (C) The base composition for SNPs on the hyper- (left) or hypo- (right) methylated alleles near all sequence-dependent ASM sites is shown as a sequence logo. (D) The median methylation levels of various 4-mer CG motifs (“Observed mCG/CG”) across the frontal cortex genome (excluding the promoters and CGIs) are shown. Motifs that contain GCG/CGC or ACG/CGT signatures are in red or blue, respectively. Examples of motif pairs (marked by “*” or “#”) with similar GC content but with distinct methylation levels are indicated. (E) The percentages of occurrences on hyper- (red) and hypo- (green) methylated alleles are shown for each 6-mer CG motif. (F) The number of total occurrences on both alleles (blue bars, with the scale at the bottom) and the p-value (binomial test, after Bonferroni multiple test correction) reflecting allele occurrence bias (red bars, with the scale at the top) for each motif in (E) are shown. (G) A scatter plot is shown for various 6-mer CG motifs comparing the Methylation Indexes to the median methylation levels across the frontal cortex genome. Motifs with tandem CGs (blue) or GCG/CGC signatures (red) are indicated. R, the Pearson correlation coefficient. (H) A similar plot as (G) is shown for various 6-mer CG motifs comparing the Methylation Indexes derived from mice to the median methylation levels across the genome of IMR90 (Lister et al., 2011). (I) The Pearson correlation coefficients are shown comparing the Methylation Indexes derived from mice to the median methylation levels across the genomes of 14 human lines (Lister et al., 2011) for various 6-mer CG motifs. See also Figure S7.
