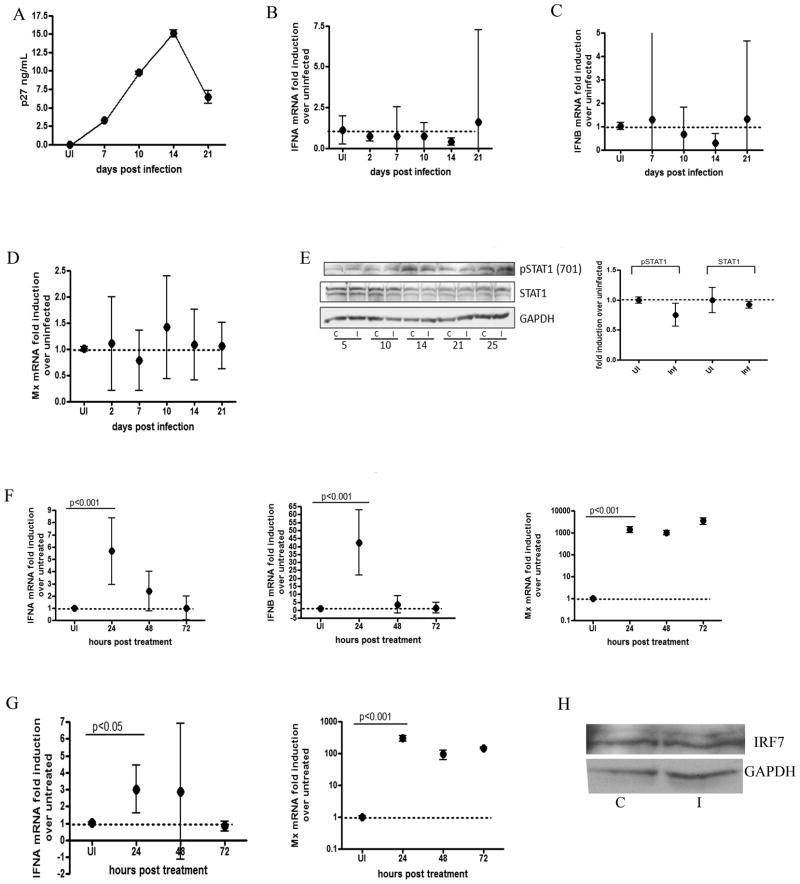
Figure 2. Type I IFN signaling is down regulated in SIV infected astrocytes.
Primary rhesus astrocytes were infected with SIV. RNA was isolated at 2, 7, 10, 14 and 21 days p.i. (A) p27 levels were measured in the supernatants by ELISA. (B–D) qRT-PCR analysis was done for (B) IFNα, (C) IFNβ and (D) MxA. TRAIL mRNA was undetectable. All qRT-PCR data is expressed as a fold induction over uninfected controls. All genes are normalized to 18s rRNA. Data is expressed as the mean of 3 independent experiments (each done with duplicate technical replicates) with 95% confidence interval. (E) Protein lysates were made at 5, 10, 14, 21 and 25 days p.i. from uninfected controls (C) and SIV infected (I) primary rhesus astrocytes and probed for GAPDH, STAT1, and pSTAT1 (Y701) via western blot. Quantitations were performed on 4 replicate western blots at 14 days p.i normalized to GAPDH. Blot shown is representative of all experiments. (F) Astrocytes were stimulated with polyI:C (10ug/mL) and RNA was isolated at 24, 28 and 72 hours p.i. qRT-PCR was done on IFNα, IFNβ and Mx. Values are normalized to 18s rRNA and are expressed as a fold induction over untreated controls. (G) Primary rhesus astrocytes were treated with IFNB (100 U/mL) and RNA was isolated 24, 48 and 72 hours p.i. qRT-PCR was done for IFNA and Mx. Values are normalized to 18s rRNA and are expressed as a fold induction over untreated controls. Data for astrocyte treatments is expressed as the means of duplicate independent experiments (each done with 3 technical replicates) with 95% confidence interval. (H) Western blot for IRF7 and GAPDH were performed on control (c) and infected (I) astrocytes 14 days p.i. Blot is representative of 3 replicates. For all mRNA comparisons, one-way ANOVA with Bonferroni post test for multiple comparisons was performed. Significant (p<0.05) pairwise comparisons are shown.