Abstract
A protein homologous to SRP54, a subunit of the mammalian signal recognition particle (SRP), was identified in Mycoplasma mycoides. The mycoplasma protein was expressed in E.coli and purified to near homogeneity. It was shown to bind specifically in vitro to a small mycoplasma RNA with structural features related to the RNA component of SRP. These findings provide evidence of a ribonucleoprotein complex in mycoplasma reminiscent of SRP. A part of the RNA was protected from ribonuclease digestion in the presence of the SRP54 homologue. The protected region contains structural elements that have been highly conserved in SRP RNAs during evolution.
Full text
PDF




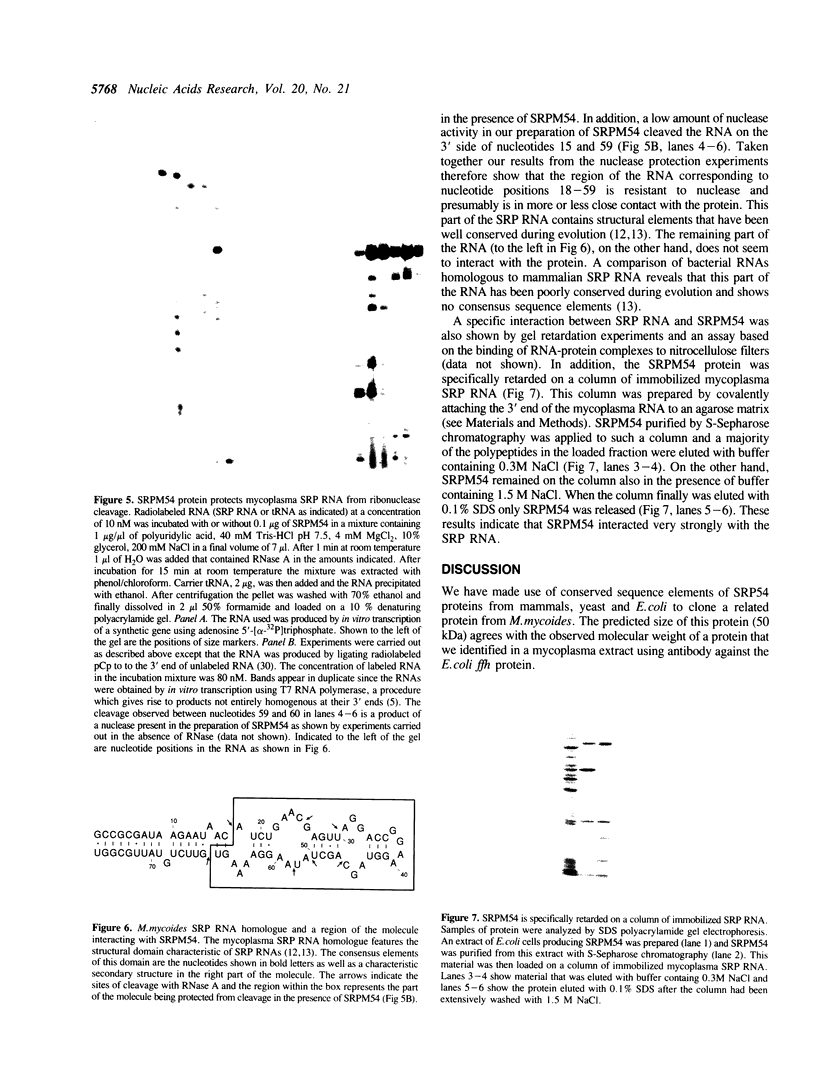


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andachi Y., Yamao F., Muto A., Osawa S. Codon recognition patterns as deduced from sequences of the complete set of transfer RNA species in Mycoplasma capricolum. Resemblance to mitochondria. J Mol Biol. 1989 Sep 5;209(1):37–54. doi: 10.1016/0022-2836(89)90168-x. [DOI] [PubMed] [Google Scholar]
- Bernstein H. D., Poritz M. A., Strub K., Hoben P. J., Brenner S., Walter P. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature. 1989 Aug 10;340(6233):482–486. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- Bourgaize D. B., Fournier M. J. Initiation of translation is impaired in E. coli cells deficient in 4.5S RNA. Nature. 1987 Jan 15;325(6101):281–284. doi: 10.1038/325281a0. [DOI] [PubMed] [Google Scholar]
- Brown S., Fournier M. J. The 4.5 S RNA gene of Escherichia coli is essential for cell growth. J Mol Biol. 1984 Sep 25;178(3):533–550. doi: 10.1016/0022-2836(84)90237-7. [DOI] [PubMed] [Google Scholar]
- Brown S. Genes for 7S RNAs can replace the gene for 4.5S RNA in growth of Escherichia coli. J Bacteriol. 1991 Mar;173(5):1835–1837. doi: 10.1128/jb.173.5.1835-1837.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S., Thon G., Tolentino E. Genetic selection and DNA sequences of 4.5S RNA homologs. J Bacteriol. 1989 Dec;171(12):6517–6520. doi: 10.1128/jb.171.12.6517-6520.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. Time of action of 4.5 S RNA in Escherichia coli translation. J Mol Biol. 1989 Sep 5;209(1):79–90. doi: 10.1016/0022-2836(89)90171-x. [DOI] [PubMed] [Google Scholar]
- Burrell H. R., Horowitz J. Affinity binding of Escherichia coli ribosomal proteins to immobilized RNA. FEBS Lett. 1975 Jan 1;49(3):306–309. doi: 10.1016/0014-5793(75)80772-1. [DOI] [PubMed] [Google Scholar]
- Byström A. S., Hjalmarsson K. J., Wikström P. M., Björk G. R. The nucleotide sequence of an Escherichia coli operon containing genes for the tRNA(m1G)methyltransferase, the ribosomal proteins S16 and L19 and a 21-K polypeptide. EMBO J. 1983;2(6):899–905. doi: 10.1002/j.1460-2075.1983.tb01519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever T. E., Glynias M. J., Merrick W. C. GTP-binding domain: three consensus sequence elements with distinct spacing. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1814–1818. doi: 10.1073/pnas.84.7.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias P., Gustafsson C. M., Hammarsten O. The origin binding protein of herpes simplex virus 1 binds cooperatively to the viral origin of replication oris. J Biol Chem. 1990 Oct 5;265(28):17167–17173. [PubMed] [Google Scholar]
- England T. E., Bruce A. G., Uhlenbeck O. C. Specific labeling of 3' termini of RNA with T4 RNA ligase. Methods Enzymol. 1980;65(1):65–74. doi: 10.1016/s0076-6879(80)65011-3. [DOI] [PubMed] [Google Scholar]
- Guindy Y. S., Samuelsson T., Johansen T. I. Unconventional codon reading by Mycoplasma mycoides tRNAs as revealed by partial sequence analysis. Biochem J. 1989 Mar 15;258(3):869–873. doi: 10.1042/bj2580869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann B. C., Poritz M. A., Walter P. Saccharomyces cerevisiae and Schizosaccharomyces pombe contain a homologue to the 54-kD subunit of the signal recognition particle that in S. cerevisiae is essential for growth. J Cell Biol. 1989 Dec;109(6 Pt 2):3223–3230. doi: 10.1083/jcb.109.6.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann B. C., Walter P. The signal recognition particle in S. cerevisiae. Cell. 1991 Oct 4;67(1):131–144. doi: 10.1016/0092-8674(91)90577-l. [DOI] [PubMed] [Google Scholar]
- High S., Dobberstein B. The signal sequence interacts with the methionine-rich domain of the 54-kD protein of signal recognition particle. J Cell Biol. 1991 Apr;113(2):229–233. doi: 10.1083/jcb.113.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu L. M., Zagorski J., Fournier M. J. Cloning and sequence analysis of the Escherichia coli 4.5 S RNA gene. J Mol Biol. 1984 Sep 25;178(3):509–531. doi: 10.1016/0022-2836(84)90236-5. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen N., Zwieb C. SRP-RNA sequence alignment and secondary structure. Nucleic Acids Res. 1991 Jan 25;19(2):209–215. doi: 10.1093/nar/19.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poritz M. A., Bernstein H. D., Strub K., Zopf D., Wilhelm H., Walter P. An E. coli ribonucleoprotein containing 4.5S RNA resembles mammalian signal recognition particle. Science. 1990 Nov 23;250(4984):1111–1117. doi: 10.1126/science.1701272. [DOI] [PubMed] [Google Scholar]
- Poritz M. A., Strub K., Walter P. Human SRP RNA and E. coli 4.5S RNA contain a highly homologous structural domain. Cell. 1988 Oct 7;55(1):4–6. doi: 10.1016/0092-8674(88)90003-7. [DOI] [PubMed] [Google Scholar]
- Razin S. Molecular biology and genetics of mycoplasmas (Mollicutes). Microbiol Rev. 1985 Dec;49(4):419–455. doi: 10.1128/mr.49.4.419-455.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy P., Birmelé C., Ebel J. P. Purification of yeast phenylalanyl-tRNA synthetase by affinity chromatography, on a tRNA(Phe)-sepharose column. FEBS Lett. 1972 Oct 15;27(1):134–138. doi: 10.1016/0014-5793(72)80426-5. [DOI] [PubMed] [Google Scholar]
- Ribes V., Römisch K., Giner A., Dobberstein B., Tollervey D. E. coli 4.5S RNA is part of a ribonucleoprotein particle that has properties related to signal recognition particle. Cell. 1990 Nov 2;63(3):591–600. doi: 10.1016/0092-8674(90)90454-m. [DOI] [PubMed] [Google Scholar]
- Römisch K., Webb J., Herz J., Prehn S., Frank R., Vingron M., Dobberstein B. Homology of 54K protein of signal-recognition particle, docking protein and two E. coli proteins with putative GTP-binding domains. Nature. 1989 Aug 10;340(6233):478–482. doi: 10.1038/340478a0. [DOI] [PubMed] [Google Scholar]
- Römisch K., Webb J., Lingelbach K., Gausepohl H., Dobberstein B. The 54-kD protein of signal recognition particle contains a methionine-rich RNA binding domain. J Cell Biol. 1990 Nov;111(5 Pt 1):1793–1802. doi: 10.1083/jcb.111.5.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelsson T., Borén T., Johansen T. I., Lustig F. Properties of a transfer RNA lacking modified nucleosides. J Biol Chem. 1988 Sep 25;263(27):13692–13699. [PubMed] [Google Scholar]
- Samuelsson T., Elias P., Lustig F., Guindy Y. S. Cloning and nucleotide sequence analysis of transfer RNA genes from Mycoplasma mycoides. Biochem J. 1985 Nov 15;232(1):223–228. doi: 10.1042/bj2320223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelsson T., Guindy Y. S., Lustig F., Borén T., Lagerkvist U. Apparent lack of discrimination in the reading of certain codons in Mycoplasma mycoides. Proc Natl Acad Sci U S A. 1987 May;84(10):3166–3170. doi: 10.1073/pnas.84.10.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelsson T., Guindy Y. Nucleotide sequence of a Mycoplasma mycoides RNA which is homologous to E. coli 4.5S RNA. Nucleic Acids Res. 1990 Aug 25;18(16):4938–4938. doi: 10.1093/nar/18.16.4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelsson T., Olsson M. Transfer RNA pseudouridine synthases in Saccharomyces cerevisiae. J Biol Chem. 1990 May 25;265(15):8782–8787. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Walter P., Lingappa V. R. Mechanism of protein translocation across the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1986;2:499–516. doi: 10.1146/annurev.cb.02.110186.002435. [DOI] [PubMed] [Google Scholar]
- Yamao F., Muto A., Kawauchi Y., Iwami M., Iwagami S., Azumi Y., Osawa S. UGA is read as tryptophan in Mycoplasma capricolum. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2306–2309. doi: 10.1073/pnas.82.8.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zopf D., Bernstein H. D., Johnson A. E., Walter P. The methionine-rich domain of the 54 kd protein subunit of the signal recognition particle contains an RNA binding site and can be crosslinked to a signal sequence. EMBO J. 1990 Dec;9(13):4511–4517. doi: 10.1002/j.1460-2075.1990.tb07902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]