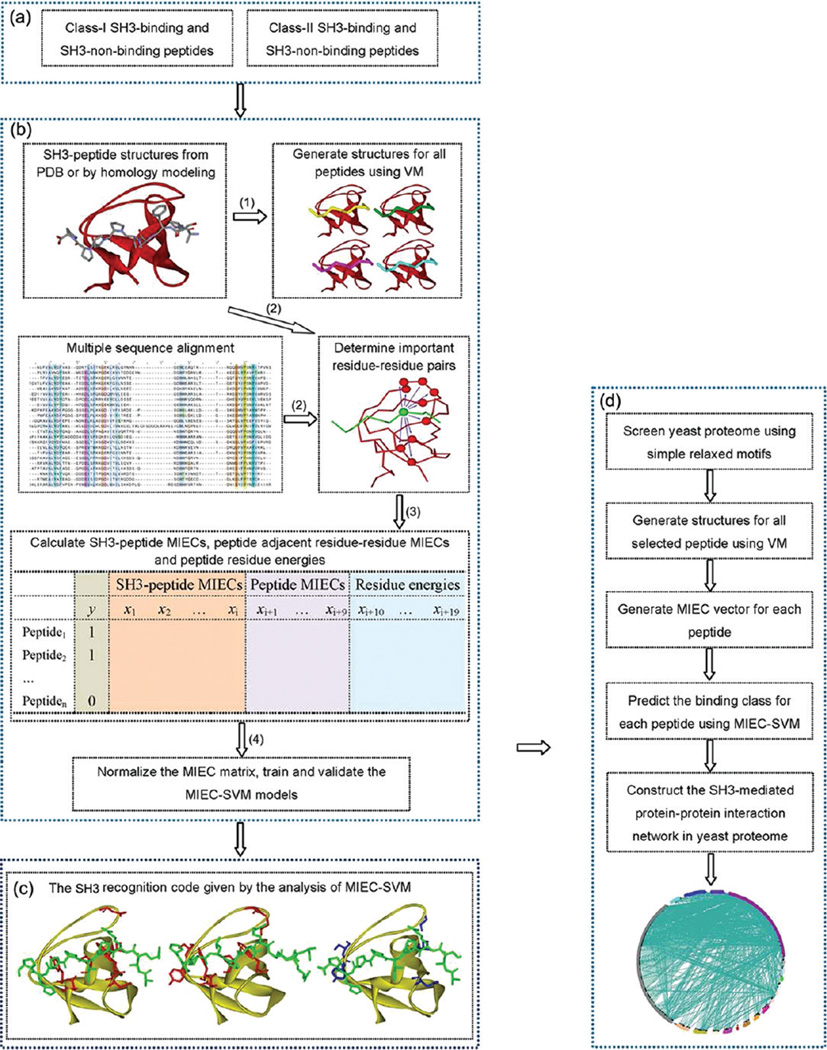
Figure 2.
Determination of the SH3 recognition codes and the predictions of the SH3-mediated protein–protein interaction network using the MIEC-SVM models. (a) Peptide binders for two classes of SH3 were collected from literature; some peptide nonbinders were collected from literature and the others were randomly selected from the Swissprot sequence database with the predefined motifs. (b) Procedure to construct the MIEC-SVM models: (1). Model the SH3-peptide complexes using Virtual Mutagenesis (VM) and GB-based molecular mechanics minimization; (2) Identify the important SH3 residues that form effective interactions with the peptides based on the complex structures and the multiple sequence alignment (the residue of peptide is shown as the green CPK model and the SH3 residues which can form effective interactions with the peptide residue are shown as the red CPK models); (3) Calculate the SH3-peptide MIECs, the peptide adjacent residue–residue MIECs and the peptide residue energies using the MM/GBSA free energy decomposition analysis; the calculation results are saved as a MIEC matrix; In the MIEC matrix, column y is the binding class for each peptide, 1 for binder and −1 for nonbinder; columns x1 to xi are the MIECs for the SH3-peptide interaction pairs; columns xi+1 to xi+9 are the MIECs for the nine pairs between the adjacent peptide residues; columns xi+10 to xi+19 are the energies for the ten residues in a peptide; it should be noted in this figure only one energy term is used; (4) Normalize the MIEC matrix, train and validate the universal MIEC-SVM models. (c) Determination of the SH3 recognition codes by analysis of the MIEC-SVM models; as an example, the important residues of the Lsb3 SH3 for recognizing the class-I peptide binding, the important residues of the Lsb3 SH3 for recognizing the class-II peptide binding, and the different residues of the Lsb3 SH3 for these two different classes of peptides are shown in three figures from left to right. (d). Construction of the SH3-mediated protein–protein interaction network in the yeast proteome by the two universal MIEC-SVM models.