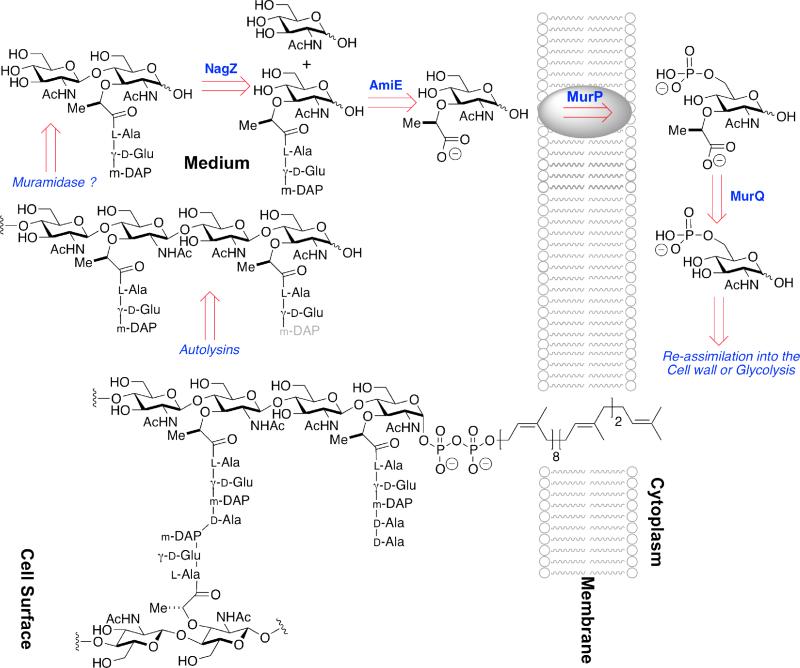
Scheme 3.
A schematic overview of MurNAc recycling in the Gram-positive monoderm B. subtilis. The single membrane of the bacterium is depicted as a cartoon running vertically to the right of center. The cytoplasm of the bacterium is found to the right of the membrane, while the cell surface is found to the left. In Gram-positive bacteria the cell surface consists of thick (relative to Gram-negative bacteria) peptidoglycan (in the example of B. subtilis, approximately 35 nm),36 to which proteins are both covalently and non-covalently attached. The membrane-bound peptidoglycan-lipid (lower structure) represents a biosynthetic intermediate, here shown with one uncross-linked peptide stem and with one cross-linked (to an adjacent glycan strand) peptide stem. Autolysin (including a not-yet-characterized muramidase) releases the GlcNAc-MurNAc disaccharide (upper left). NagZ-dependent glycosylase activity splits the disaccharide, following which AmiE (possibly in concert with other amidases) releases the peptide. MurNAc is internalized by the MurP phosphotransferase, after which the lactyl ether is cleaved by the MurQ etherase. This schematic for B. subtilis should not be taken as general: Reith and Mayer have obtained strong evidence for a wholly cytoplasmic recycling pathway in the Gram-positive bacterium Clostridium acetobutylicum.37,38