Abstract
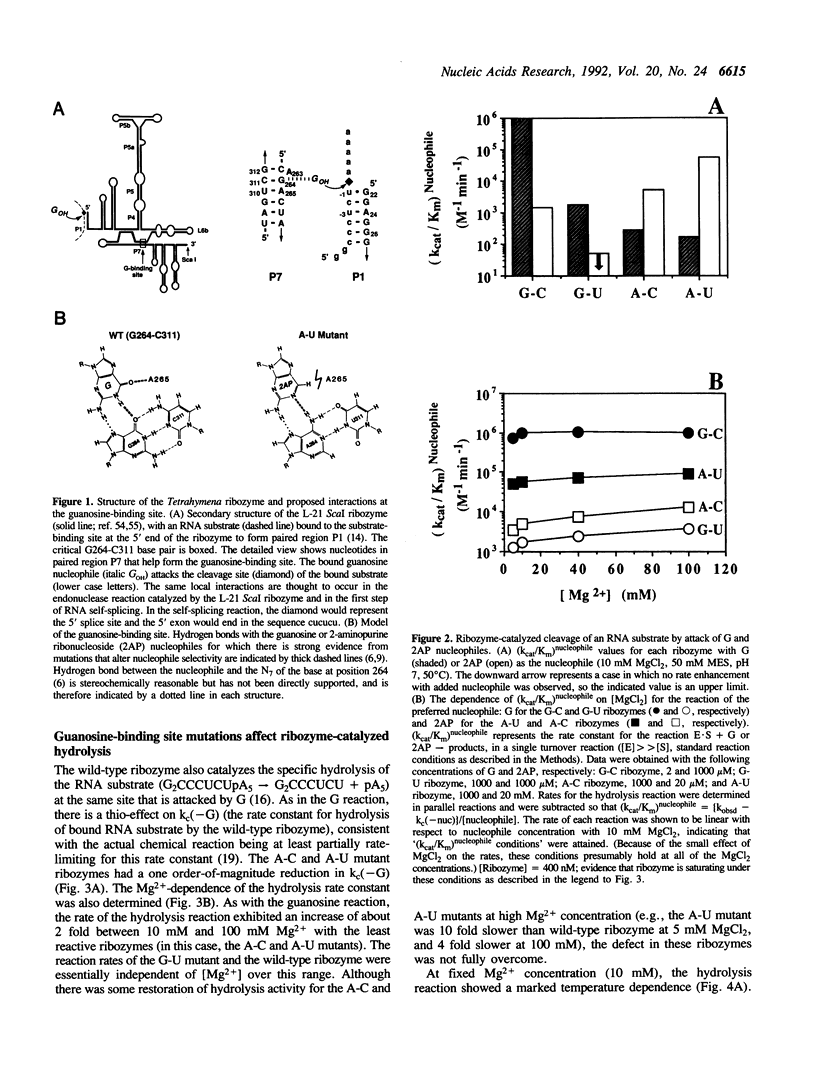
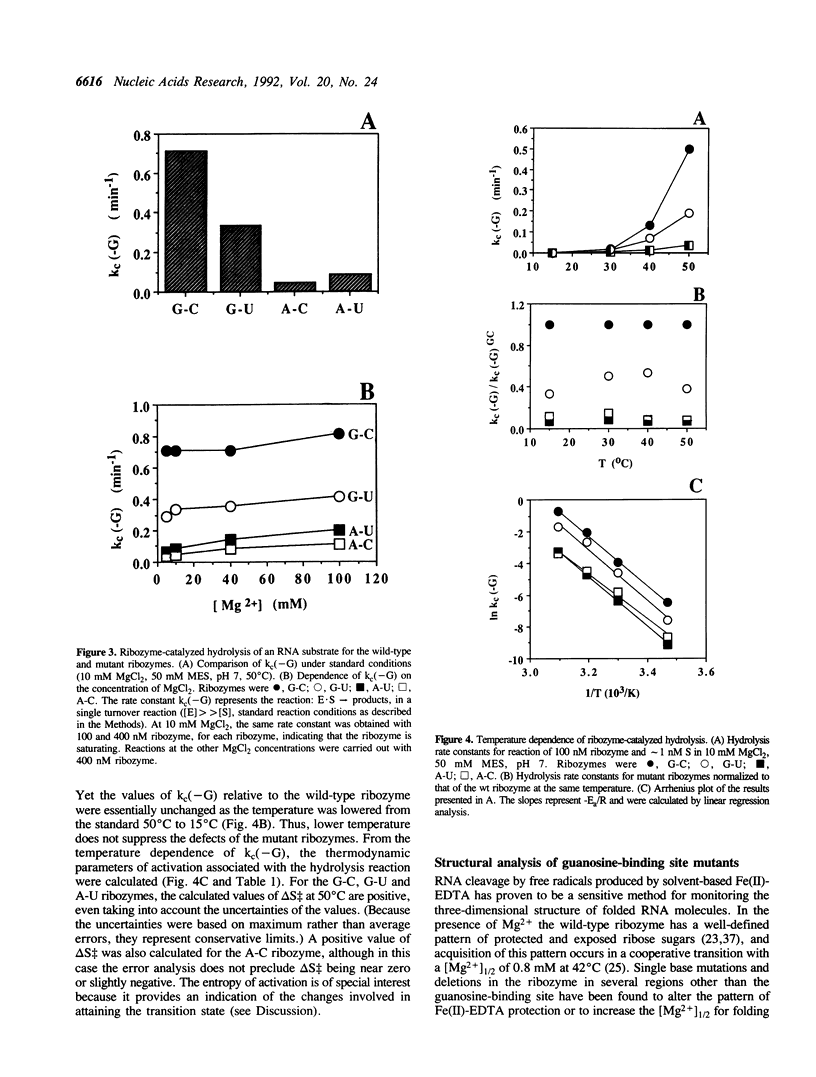
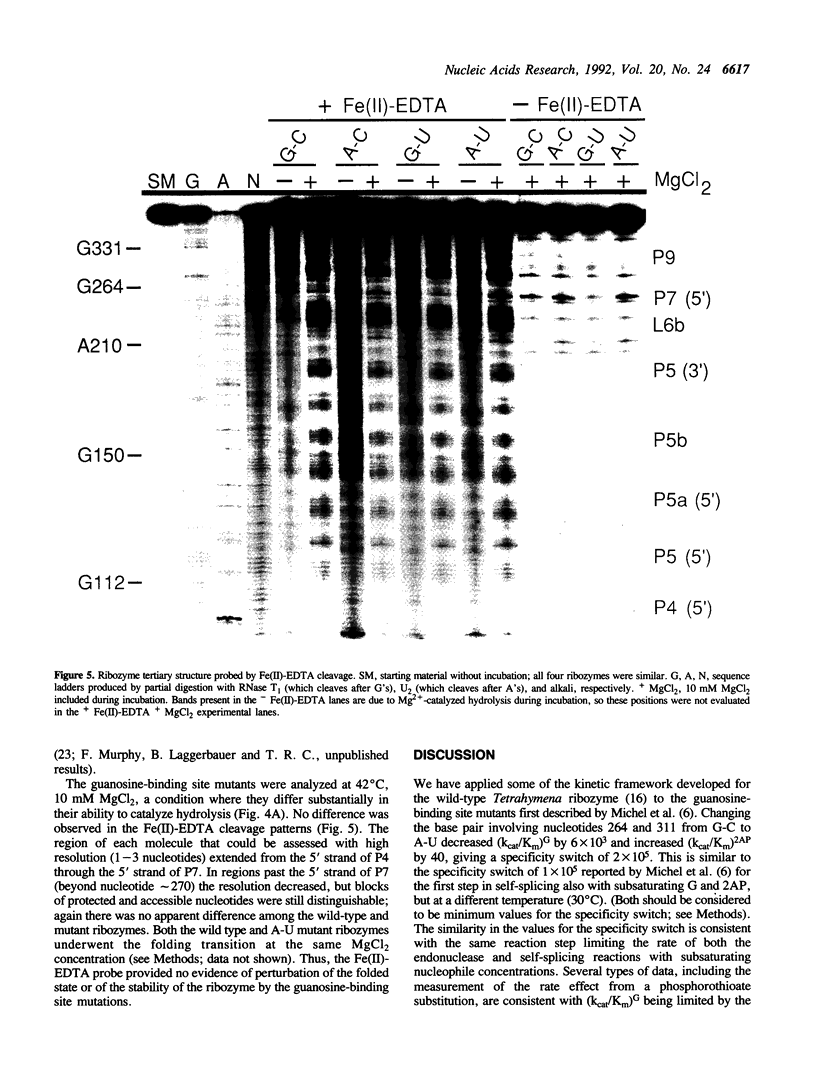
Self-splicing group I introns use guanosine as a nucleophile to cleave the 5' splice site. The guanosine-binding site has been localized to the G264-C311 base pair of the Tetrahymena intron on the basis of analysis of mutations that change the specificity of the nucleophile from G (guanosine) to 2AP (2-aminopurine ribonucleoside) (F. Michel et al. (1989) Nature 342, 391-395). We studied the effect of these mutations (G-U, A-C and A-U replacing G264-C311) in the L-21 ScaI version of the Tetrahymena ribozyme. In this enzymatic system (kcat/Km)G monitors the cleavage step. This kinetic parameter decreased by at least 5 x 10(3) when the G264-C311 base pair was mutated to an A-U pair, while (kcat/Km)2AP increased at least 40-fold. This amounted to an overall switch in specificity of at least 2 x 10(5). The nucleophile specificity (G > 2AP for the G-C and G-U pairs, 2AP > G for the A-U and A-C pairs) was consistent with the proposed hydrogen bond between the nucleotide at position 264 and N1 of the nucleophile. Unexpectedly, the A-U and A-C mutants showed a decrease of an order of magnitude in the rate of ribozyme-catalyzed hydrolysis of RNA, in which H2O or OH- replaces G as the nucleophile, whereas the G-U mutant showed a decrease of only 2-fold. The low hydrolysis rates were not restored by raising the Mg2+ concentration or lowering the temperature. In addition, the mutant ribozymes exhibited a pattern of cleavage by Fe(II)-EDTA indistinguishable from that of the wild type, and the [Mg2+]1/2 for folding of the A-U mutant ribozyme was the same as that of the wild type. Therefore the guanosine-binding site mutations do not appear to have a major effect on RNA folding or stability. Because changing G264 affects the hydrolysis reaction without perturbing the global folding of the RNA, we conclude that the catalytic role of this conserved nucleotide is not limited to guanosine binding.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bass B. L., Cech T. R. Ribozyme inhibitors: deoxyguanosine and dideoxyguanosine are competitive inhibitors of self-splicing of the Tetrahymena ribosomal ribonucleic acid precursor. Biochemistry. 1986 Aug 12;25(16):4473–4477. doi: 10.1021/bi00364a001. [DOI] [PubMed] [Google Scholar]
- Bass B. L., Cech T. R. Specific interaction between the self-splicing RNA of Tetrahymena and its guanosine substrate: implications for biological catalysis by RNA. 1984 Apr 26-May 2Nature. 308(5962):820–826. doi: 10.1038/308820a0. [DOI] [PubMed] [Google Scholar]
- Been M. D., Perrotta A. T. Group I intron self-splicing with adenosine: evidence for a single nucleoside-binding site. Science. 1991 Apr 19;252(5004):434–437. doi: 10.1126/science.2017681. [DOI] [PubMed] [Google Scholar]
- Burke J. M., Irvine K. D., Kaneko K. J., Kerker B. J., Oettgen A. B., Tierney W. M., Williamson C. L., Zaug A. J., Cech T. R. Role of conserved sequence elements 9L and 2 in self-splicing of the Tetrahymena ribosomal RNA precursor. Cell. 1986 Apr 25;45(2):167–176. doi: 10.1016/0092-8674(86)90380-6. [DOI] [PubMed] [Google Scholar]
- Cech T. R. Conserved sequences and structures of group I introns: building an active site for RNA catalysis--a review. Gene. 1988 Dec 20;73(2):259–271. doi: 10.1016/0378-1119(88)90492-1. [DOI] [PubMed] [Google Scholar]
- Cech T. R. Self-splicing of group I introns. Annu Rev Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Zaug A. J., Grabowski P. J. In vitro splicing of the ribosomal RNA precursor of Tetrahymena: involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell. 1981 Dec;27(3 Pt 2):487–496. doi: 10.1016/0092-8674(81)90390-1. [DOI] [PubMed] [Google Scholar]
- Celander D. W., Cech T. R. Iron(II)-ethylenediaminetetraacetic acid catalyzed cleavage of RNA and DNA oligonucleotides: similar reactivity toward single- and double-stranded forms. Biochemistry. 1990 Feb 13;29(6):1355–1361. doi: 10.1021/bi00458a001. [DOI] [PubMed] [Google Scholar]
- Celander D. W., Cech T. R. Visualizing the higher order folding of a catalytic RNA molecule. Science. 1991 Jan 25;251(4992):401–407. doi: 10.1126/science.1989074. [DOI] [PubMed] [Google Scholar]
- Del Sal G., Manfioletti G., Schneider C. A one-tube plasmid DNA mini-preparation suitable for sequencing. Nucleic Acids Res. 1988 Oct 25;16(20):9878–9878. doi: 10.1093/nar/16.20.9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor P. J., Flanegan J. B., Cech T. R. A conserved base pair within helix P4 of the Tetrahymena ribozyme helps to form the tertiary structure required for self-splicing. EMBO J. 1989 Nov;8(11):3391–3399. doi: 10.1002/j.1460-2075.1989.tb08503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampel A., Cech T. R. Binding of the CBP2 protein to a yeast mitochondrial group I intron requires the catalytic core of the RNA. Genes Dev. 1991 Oct;5(10):1870–1880. doi: 10.1101/gad.5.10.1870. [DOI] [PubMed] [Google Scholar]
- Good N. E., Winget G. D., Winter W., Connolly T. N., Izawa S., Singh R. M. Hydrogen ion buffers for biological research. Biochemistry. 1966 Feb;5(2):467–477. doi: 10.1021/bi00866a011. [DOI] [PubMed] [Google Scholar]
- Grosshans C. A., Cech T. R. A hammerhead ribozyme allows synthesis of a new form of the Tetrahymena ribozyme homogeneous in length with a 3' end blocked for transesterification. Nucleic Acids Res. 1991 Jul 25;19(14):3875–3880. doi: 10.1093/nar/19.14.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschlag D., Cech T. R. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 1. Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry. 1990 Nov 6;29(44):10159–10171. doi: 10.1021/bi00496a003. [DOI] [PubMed] [Google Scholar]
- Herschlag D., Cech T. R. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 2. Kinetic description of the reaction of an RNA substrate that forms a mismatch at the active site. Biochemistry. 1990 Nov 6;29(44):10172–10180. doi: 10.1021/bi00496a004. [DOI] [PubMed] [Google Scholar]
- Herschlag D., Cech T. R. DNA cleavage catalysed by the ribozyme from Tetrahymena. Nature. 1990 Mar 29;344(6265):405–409. doi: 10.1038/344405a0. [DOI] [PubMed] [Google Scholar]
- Herschlag D., Piccirilli J. A., Cech T. R. Ribozyme-catalyzed and nonenzymatic reactions of phosphate diesters: rate effects upon substitution of sulfur for a nonbridging phosphoryl oxygen atom. Biochemistry. 1991 May 21;30(20):4844–4854. doi: 10.1021/bi00234a003. [DOI] [PubMed] [Google Scholar]
- Heuer T. S., Chandry P. S., Belfort M., Celander D. W., Cech T. R. Folding of group I introns from bacteriophage T4 involves internalization of the catalytic core. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11105–11109. doi: 10.1073/pnas.88.24.11105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Sullivan F. X., Cech T. R. New reactions of the ribosomal RNA precursor of Tetrahymena and the mechanism of self-splicing. J Mol Biol. 1986 May 5;189(1):143–165. doi: 10.1016/0022-2836(86)90387-6. [DOI] [PubMed] [Google Scholar]
- Joyce G. F., van der Horst G., Inoue T. Catalytic activity is retained in the Tetrahymena group I intron despite removal of the large extension of element P5. Nucleic Acids Res. 1989 Oct 11;17(19):7879–7889. doi: 10.1093/nar/17.19.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAUZMANN W. Some factors in the interpretation of protein denaturation. Adv Protein Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- Kay P. S., Inoue T. Catalysis of splicing-related reactions between dinucleotides by a ribozyme. 1987 May 28-Jun 3Nature. 327(6120):343–346. doi: 10.1038/327343a0. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Ladner J. E., Jack A., Robertus J. D., Brown R. S., Rhodes D., Clark B. F., Klug A. Structure of yeast phenylalanine transfer RNA at 2.5 A resolution. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4414–4418. doi: 10.1073/pnas.72.11.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham J. A., Cech T. R. Defining the inside and outside of a catalytic RNA molecule. Science. 1989 Jul 21;245(4915):276–282. doi: 10.1126/science.2501870. [DOI] [PubMed] [Google Scholar]
- McSwiggen J. A., Cech T. R. Stereochemistry of RNA cleavage by the Tetrahymena ribozyme and evidence that the chemical step is not rate-limiting. Science. 1989 May 12;244(4905):679–683. doi: 10.1126/science.2470150. [DOI] [PubMed] [Google Scholar]
- Michel F., Dujon B. Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J. 1983;2(1):33–38. doi: 10.1002/j.1460-2075.1983.tb01376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Hanna M., Green R., Bartel D. P., Szostak J. W. The guanosine binding site of the Tetrahymena ribozyme. Nature. 1989 Nov 23;342(6248):391–395. doi: 10.1038/342391a0. [DOI] [PubMed] [Google Scholar]
- Michel F., Jaeger L., Westhof E., Kuras R., Tihy F., Xu M. Q., Shub D. A. Activation of the catalytic core of a group I intron by a remote 3' splice junction. Genes Dev. 1992 Aug;6(8):1373–1385. doi: 10.1101/gad.6.8.1373. [DOI] [PubMed] [Google Scholar]
- Michel F., Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990 Dec 5;216(3):585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987 Nov 11;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle A. M., Cech T. R. Ribozyme recognition of RNA by tertiary interactions with specific ribose 2'-OH groups. Nature. 1991 Apr 18;350(6319):628–631. doi: 10.1038/350628a0. [DOI] [PubMed] [Google Scholar]
- Pyle A. M., McSwiggen J. A., Cech T. R. Direct measurement of oligonucleotide substrate binding to wild-type and mutant ribozymes from Tetrahymena. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8187–8191. doi: 10.1073/pnas.87.21.8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle A. M., Murphy F. L., Cech T. R. RNA substrate binding site in the catalytic core of the Tetrahymena ribozyme. Nature. 1992 Jul 9;358(6382):123–128. doi: 10.1038/358123a0. [DOI] [PubMed] [Google Scholar]
- Quigley G. J., Rich A. Structural domains of transfer RNA molecules. Science. 1976 Nov 19;194(4267):796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- Rajagopal J., Doudna J. A., Szostak J. W. Stereochemical course of catalysis by the Tetrahymena ribozyme. Science. 1989 May 12;244(4905):692–694. doi: 10.1126/science.2470151. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto N., Kierzek R., Turner D. H. Kinetics for reaction of a circularized intervening sequence with CU, UCU, CUCU, and CUCUCU: mechanistic implications from the dependence on temperature and on oligomer and Mg2+ concentrations. Biochemistry. 1988 Aug 23;27(17):6384–6392. doi: 10.1021/bi00417a029. [DOI] [PubMed] [Google Scholar]
- Sugimoto N., Tomka M., Kierzek R., Bevilacqua P. C., Turner D. H. Effects of substrate structure on the kinetics of circle opening reactions of the self-splicing intervening sequence from Tetrahymena thermophila: evidence for substrate and Mg2+ binding interactions. Nucleic Acids Res. 1989 Jan 11;17(1):355–371. doi: 10.1093/nar/17.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner N. K., Cech T. R. Guanosine binding required for cyclization of the self-splicing intervening sequence ribonucleic acid from Tetrahymena thermophila. Biochemistry. 1987 Jun 16;26(12):3330–3340. doi: 10.1021/bi00386a013. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Iron(II) EDTA used to measure the helical twist along any DNA molecule. Science. 1985 Nov 8;230(4726):679–681. doi: 10.1126/science.2996145. [DOI] [PubMed] [Google Scholar]
- Waring R. B., Scazzocchio C., Brown T. A., Davies R. W. Close relationship between certain nuclear and mitochondrial introns. Implications for the mechanism of RNA splicing. J Mol Biol. 1983 Jul 5;167(3):595–605. doi: 10.1016/s0022-2836(83)80100-4. [DOI] [PubMed] [Google Scholar]
- Yarus M., Illangesekare M., Christian E. An axial binding site in the Tetrahymena precursor RNA. J Mol Biol. 1991 Dec 20;222(4):995–1012. doi: 10.1016/0022-2836(91)90590-3. [DOI] [PubMed] [Google Scholar]
- Yarus M., Majerfeld I. Co-optimization of ribozyme substrate stacking and L-arginine binding. J Mol Biol. 1992 Jun 20;225(4):945–949. doi: 10.1016/0022-2836(92)90095-2. [DOI] [PubMed] [Google Scholar]
- Young B., Herschlag D., Cech T. R. Mutations in a nonconserved sequence of the Tetrahymena ribozyme increase activity and specificity. Cell. 1991 Nov 29;67(5):1007–1019. doi: 10.1016/0092-8674(91)90373-7. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Been M. D., Cech T. R. The Tetrahymena ribozyme acts like an RNA restriction endonuclease. Nature. 1986 Dec 4;324(6096):429–433. doi: 10.1038/324429a0. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Grosshans C. A., Cech T. R. Sequence-specific endoribonuclease activity of the Tetrahymena ribozyme: enhanced cleavage of certain oligonucleotide substrates that form mismatched ribozyme-substrate complexes. Biochemistry. 1988 Dec 13;27(25):8924–8931. doi: 10.1021/bi00425a008. [DOI] [PubMed] [Google Scholar]