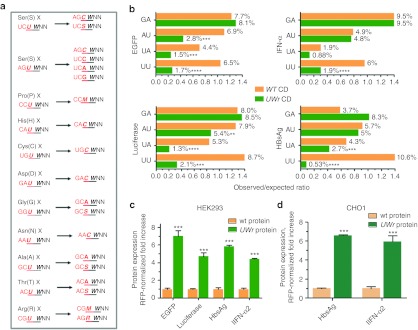
Figure 1.
Impact of UW dinucleotide frequency on protein expression. (a) The dicodon change algorithm eliminates UU or UA at the boundary by replacing the first amino acid codon that ends with U with a synonymous codon that ends with an S nucleotide. X is any amino acid. S (G or C), M (A or C), R (A or G) nucleotides are IUPAC symbols. (b) Dinucleotide frequency analysis in WT and UW reduced mRNA-coding region constructs. The Compseq program, an EMBOSS algorithm29 that calculates nucleotide composition in a sequence, was used http://mobyle.pasteur.fr/cgi-bin/portal.py#forms::compseq. The expected frequency of any dinucleotide is 1/16 (6.25%). Thus, the observed to expected ratio = % observed dinucleotide/6.25%. The observed dinucleotide (%) is shown on the columns. Several coding regions were designed based on the algorithm shown in figure. These were custom synthesized, and then subcloned into expression vectors. Two cell lines (c) HEK293 or (d) CHO1 were cotransfected with expression vectors containing wild-type (WT) coding region or UW-reduced coding region (UWr) of different genes and red fluorescent protein (RFP) plasmid at a ratio of 1/4. Protein expression at 18 hours after transfection was quantified by fluorescence (EGFP), chemiluminescence (luciferase), or enzyme-linked immunosorbent assay (ELISA) [interferon-α (IFN-α) and hepatitis B surface antigen (HBsAg)]. Data are presented as RFP-normalized fold increase which = (Expression values of UWr plasmid/WT plasmid) × (RFP normalization ratio). Data are mean ±SEM of three independent experiments.