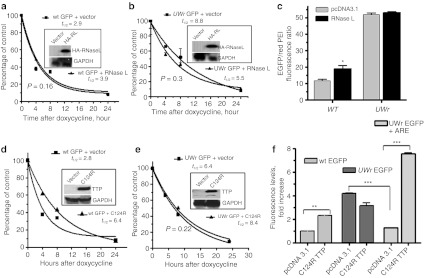
Figure 4.
Response of the enhanced green fluorescent protein (EGFP)-coding region variants to RNase L and dominant negative zinc-finger mutant, C124R tristetraprolin (TTP). HeLa Tet-off cell cells (3 × 104 cells/well) in 96-well microplates were cotransfected with expression vectors containing (a) wild-type (WT) coding region or (b) UW-reduced coding region (UWr) for EGFP mRNA and each of RNase L or pcDNA3.1 control expression vector. Subsequently, the transcription from the cotransfected EGFP plasmid was blocked by doxycycline (1 µg/ml) for the indicated periods of time. Total RNA was extracted and subjected in reverse transcription (RT)-QPCR using TaqMan primers specific to WT and UWr EGFP mRNA (details in Materials and Methods). The one-phase exponential mRNA decay model7 was used to quantify the mRNA half-life for WT (R2 = 0.96 and 0.99 for vector and RNase L curves) and UW-reduced EGFP mRNAs (R2 = 0.9 and 0.85, vector and RNase L, respectively). The data are mean ± SEM of replicate from one representative experiment of two. Insets show Western blotting for transfected HA-tagged RNase L (HA-RL) expression using a specific antibody to HA tag. (c) Protein expression changes as a result of RNase L expression using fluorescence level ratio of green fluorescence to red fluorescence. Details of experiment are similar to those described in legend A and B. (d,e) HeLa Tet-off cells (3 × 104 cells/well) in 96-well microplates were transfected for 18 hours with expression vectors containing (d) wild-type (WT) coding region or (e) UWr-coding region for EGFP in the presence of C124R expression vector or pcDNA3.1 control vector. Subsequently, the transcription was blocked by doxycycline (1 µg/ml) for the indicated periods of time. The one-phase exponential mRNA decay model7 was used to quantify the mRNA half life for WT (R2 = 0.97 and 0.99 for vector and C124R, respectively) and UWr EGFP mRNAs (R2 = 0.96 and 0.9.5 for vector and C124R, respectively). The data are mean ± SEM of three replicates from one representative experiment of two independent experiments performed. Insets show Western blotting for C124R expression. (f) Protein expression from WT EGFP-coding region, UWr EGFP-coding region, and UWr EGFP-coding region fused with tumor necrosis factor (TNF)-α ARE 3′UTR. Data is presented as fluorescence levels (mean ± SEM) of three independent experiments, each with three to four replicates. WT EGFP + pcDNA3.1 vector control was normalized to 1.0-fold ratio. The UWr EGFP and UWr EGFP+ARE data were normalized to UWr EGFP + pcDNA 3.1 control (fold ratio = 1.0).