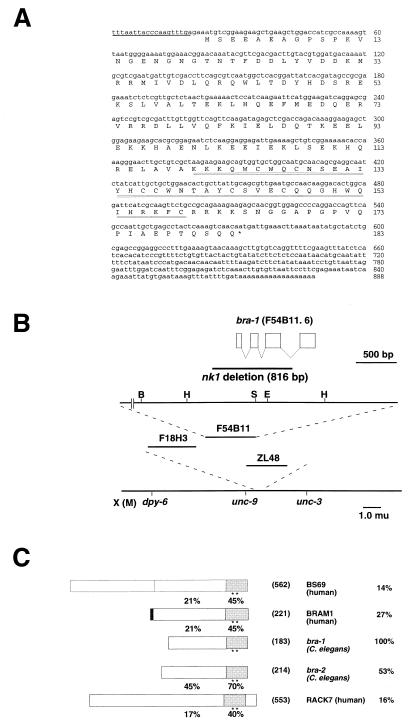
Figure 1.
Structural analysis of bra-1. (A) Nucleotide and predicted amino acid sequences of bra-1 cDNA. SL1 trans-spliced leader sequence is indicated with an underline. The conserved zinc-finger-like sequence is indicated with a double underline. (B) A physical and genetic map of the bra-1 region. The top line is the gene structure with exons shown in boxes separated by introns. The direction of transcription is from right to left. The deletion allele of the bra-1 gene was isolated by PCR screening of populations of Tc1 insertion strains for an imprecise Tc1 excision. The extent of the nk1 deletion, 816 bp, which correspond to cosmid F54B11 25420–26236, is shown. The second line is the partial restriction map of the genomic cosmid clone F54B11. B, BamHI; H, HindIII; S, SacI; E, EcoRI. The third line is the physical map showing the position of cosmid clone F54B11. The fourth line is the genetic map of a portion close to the center of the X chromosome. (C) Alignment of the predicted amino acid sequences of the structure of BS69 (human), BRAM1 (human), BRA-1 (C. elegans), BRA-2 (C. elegans), and RACK7 (human). The percent identities for the whole sequences with BRA-1 are indicated to the left. The percent identities of N-terminal and C-terminal regions are indicated next to respective regions. Conserved zinc-finger-like motifs are indicated with asterisks. Numbers of predicted amino acid residues are given in parentheses.