Abstract
Bacterial lipoproteins have many important functions owing to their essential nature and roles in pathogenesis and represent a class of possible vaccine candidates. The prediction of bacterial lipoproteins from sequence is thus an important task for computational vaccinology. A Support Vector Machines (SVM) based module for predicting bacterial lipoproteins, LIPOPREDICT, has been developed. The best performing sequence model were generated using selected dipeptide composition, which gave 97% accuracy of prediction. The results obtained were compared very well with those of previously developed methods.
Availability
The database is available for free at www.lipopredict.cdac.in
Keywords: Bacterial lipoproteins, Support Vector Machine (SVM), compositional features, prediction server
Background
Bacterial lipoproteins have many important functions owing to their essential nature and roles in pathogenesis representing a class of possible vaccine candidates. They are functionally diverse class of membrane-anchored proteins that typically represent approximately 2% of the bacterial proteome [1]. They consist of a large group of proteins and perform many different functions: promote antibiotic resistance, cell signaling and substrate binding in ABC transport systems, protein export, sporulation, germination, bacterial conjugation, and many others are yet to be assigned a function [2]. Lipoproteins are required for virulence in many bacteria. They perform variety of roles in host-pathogen interaction, from surface adhesion and initiation of inflammatory processes through translocation of virulence factors into the host cytoplasm [3]. Several methods have been devised in literature to predict bacterial lipoproteins, using different approaches and data sets. Identification of Gram-positive bacterial lipoproteins has resulted in various servers and databases such as DOLOP [4], LIPO [5], PSORT [6] and ScanProsite [7]. LipoP [8] and Phobius [9] use hidden Markov model. LipPred [10] uses Naive-Bayesian network and SPEPLip [11] uses neural network. LipoP [8] identification of Gram- negative bacterial lipoproteins uses pattern matching methodology. In this work, we present a SVM based method using amino acid composition to identify bacterial lipoproteins.
Methodology
Bacterial Lipoprotein Dataset:
The dataset of bacterial lipoproteins consists of experimentally annotated 222 sequences. It has been derived from distinct bacterial lipoproteins available in the DOLOP [4] database.
Bacterial Non Lipoprotein Dataset:
222 bacterial non lipoprotein sequences which were obtained from various databases such as NCBI [12], UNIPROT [13] were used for the construction of the dataset.
Both the datasets were compiled after performing CD-HIT. The program CD-HIT (Cluster Database at High Identity with Tolerance) [14] [15] removes homologous sequences by clustering the protein dataset at user-defined sequence identity thresholds. Here we employed multiple CD-HIT runs; for example 90%, and then 60% and then 50% generating more efficient non-redundant datasets.
Server Implementation:
The prediction method described in this paper is implemented in the form of a web-server LIPOPREDICT: Bacterial Lipoprotein prediction server (Figure 1). Bacterial lipoproteins are a diverse and functionally important group of proteins that are amenable to bioinformatic analyses because of their unique signal peptide features. They are characterized by the presence of a signal peptide in their Nterminus, followed by presence of a specific cysteine residue [16]. The lipidation motif, represented in PROSITE by the regular expression DERK (6) [LIVMFW STAG] (2) − [LIVMFYSTAGCQ] −[AGS]−C(PS00013), is present in both Gram- positive and Gram-negative bacteria. Signal peptides of bacterial lipoproteins possess many distinctive physio chemical features, along with the presence or absence of specific amino acids [17]. G. von Heijne [18] showed that considerable structural and compositional differences exist between signal peptides of bacterial lipoproteins and bacterial non lipoproteins. The two classes differ from each other in terms of the physio-chemical properties like charge, hydrophobicity, secondary structure propensities and amino acid size. All those differences in signal peptides from lipoproteins and non-lipoproteins can be attributed to the amino acid composition of the signal peptide. Hence compositional features like amino acid and dipeptide composition can be employed for discriminating these signal peptides, which in turn will result in differentiating bacterial lipoproteins and non-lipoproteins. In our work, we analyzed the amino acid sequence of the signal peptides of lipoproteins and bacterial non lipoproteins. The average length of signal peptides in bacteria ranges from 24 amino acids for Gram-negatives and 32 amino acids for Gram positives [19]. Considering few variations in the lengths, we selected first 35 residues for the analysis.
Figure 1.
Snapshot of the index page of LIPOPREDICT server.
Server description
Prediction Input Interface:
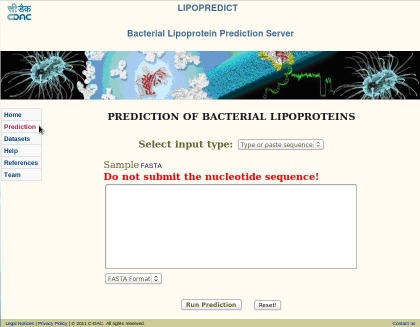
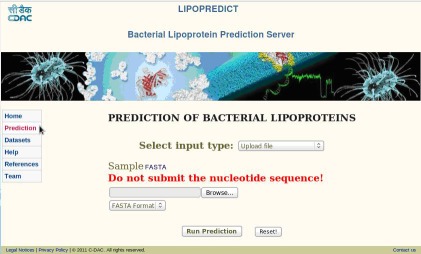
Users can click on the prediction icon and the prediction interface displays various input type option. In which user can either type or paste sequence (Figure 2) or submit the file using the option upload file (Figure 3). Submitting sequence/s must be in FASTA format, on submission the input sequence is validated and if invalid the errors are reported to the user to rectify the problem.
Figure 2.
Snapshot of query prediction page − Type or Paste Sequence.
Figure 3.
Snapshot of query prediction page − Upload File.
Prediction Output Interface:
When the run prediction button is clicked, the users are directed to the results page, best model (Selected Dipeptide Composition) with highest cross validation were created and used for prediction. Compositional feature model selected dipeptide is been used to generate prediction results. Support vector machines (SVMs) with probability estimates are calculated and the prediction result with probability estimate of the sequence belonging to the respective class is displayed in the result page. Result page also gives an option to download the prediction results for further use.
Discussion
SVM kernel types and kernel parameters were tuned based on 10 fold cross validation accuracy as performance measure. The results are tabulated in Table 1 (see supplementary material). Bacterial lipoprotein prediction problem with feature selection gave the highest accuracy of 97% with selected dipeptide composition feature. We employed information gain feature selection using WEKA software [20] with the view to extracting the subsets of informative features. With feature selection the maximum accuracy increased for selected dipeptide, so we employed 67 selected dipeptide composition features as SVM domain feature input for building the model.
Conclusions
In this study we have presented a prediction server, LIPOPREDICT for identification and classification of bacterial lipoproteins. The server employs Support Vector Machines supervisory learning model, which is rigorously based on statistical learning theory. For prediction of bacterial lipoprotein, selection of most informative features in dipeptide composition further improved model accuracy. Our results indicate that this SVM model can be employed for accurate prediction of bacterial lipoproteins. The prediction models include probability measures in the output, so it can be used to assess the confidence of SVM predictions. Further, our user friendly web server can be readily used for annotation of novel proteins.
Supplementary material
Acknowledgments
Dr. VKJ gratefully thanks CSIR, New Delhi for support in the form of Emeritus Scientist Grant. The authors also thank NPSF group members of CDAC for help and support for hosting of web server.
Footnotes
Citation:Kumari et al, Bioinformation 8(8): 394-398 (2012)
References
- 1.MI Hutchings, et al. Trends Microbiol. 2009;17:13. doi: 10.1016/j.tim.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 2.IC Sutcliffe, et al. Microbiology. 2002;148:2065. [Google Scholar]
- 3.A Kovacs-Simon, et al. Infect Immun. 2011;79:548. doi: 10.1128/IAI.00682-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.M Madan Babu, K Sankaran. Bioinformatics. 2002;18:641. doi: 10.1093/bioinformatics/18.4.641. [DOI] [PubMed] [Google Scholar]
- 5.FS Berven, et al. Arch Microbiol. 2006;184:362. doi: 10.1007/s00203-005-0055-7. [DOI] [PubMed] [Google Scholar]
- 6.K Nakai, P Horton. Trends Biochem Sci. 1999;24:34. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 7.E De Castro, et al. Nucleic Acids Res. 2006;34:W362. [Google Scholar]
- 8.AS Juncker, et al. Protein Sci. 2003;12:1652. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.L Käll, et al. J Mol Biol. 2004;338:1027. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 10.PD Taylor, et al. Bioinformation. 2006;1:176. doi: 10.6026/97320630001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.P Fariselli, et al. Bioinformatics. 2003;19:2498. [Google Scholar]
- 12. http://www.ncbi.nlm.nih.gov/
- 13. http://www.uniprot.org/
- 14.W Li, et al. Bioinformatics. 2002;18:77. doi: 10.1093/bioinformatics/18.1.77. [DOI] [PubMed] [Google Scholar]
- 15.W Li, et al. Bioinformatics. 2001;17:282. [Google Scholar]
- 16.S Hayashi, HC Wu. J Bioenerg Biomembr. 1990;22:451. doi: 10.1007/BF00763177. [DOI] [PubMed] [Google Scholar]
- 17.P Klein, et al. Protein Eng. 1988;2:15. [Google Scholar]
- 18.G Von Heijne. Protein Eng. 1989;2:531. doi: 10.1093/protein/2.7.531. [DOI] [PubMed] [Google Scholar]
- 19.JD Bendtsen, et al. J Mol Biol. 2004;340:783. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 20. http://www.cs.waikato.ac.nz/ml/weka/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.