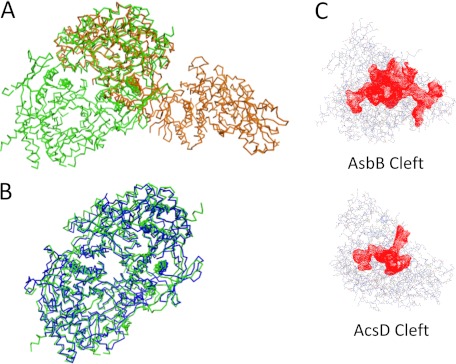
FIGURE 4.
Comparison of AsbB to other NIS synthetases. Calculations regarding structural similarity were made using PDBeFold (69). A, superposition of AsbB (green) and the type A NIS synthetase AcsD (orange) demonstrates similar monomeric structures with different dimers. Between the two proteins, the sequence identity is 20.5%, and the mean square deviation on α-carbon atom positions is 2.52 Å with 448 amino acids compared. B, superposition of AsbB (green) and the type C NIS synthetase AlcC (blue) indicates the same -fold with similar dimers. For these two proteins, the sequence identity is 24.6%, and the root mean square deviation on α-carbon atom positions is 1.79 Å with 516 amino acids compared. C, ligand binding pockets (red) of AsbB and AcsD are shown. The cleft sizes of AsbB and AcsD are 8614 and 3593 Å3, respectively, as calculated by Profunc (52, 70) and plotted by Jmol (19).