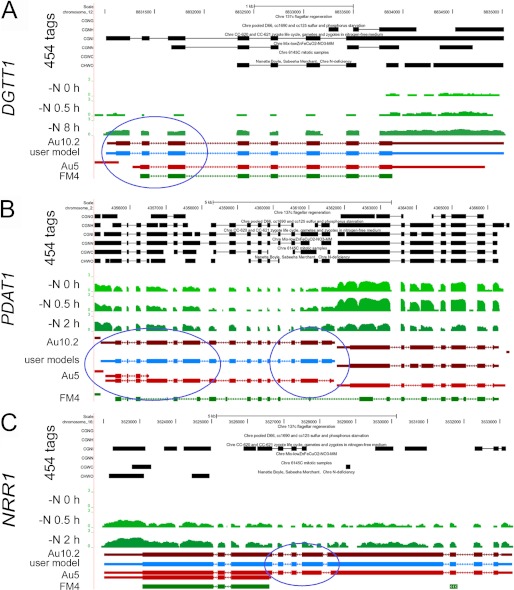
FIGURE 6.
Browser view of Chlamydomonas DGTT1 (A), PDAT1 (B), and NRR1 (C) loci. The UCSC browser was used to view RNA-Seq coverage. The genome coordinates are shown at the top. The next six tracks labeled 454 ESTs represent sequences, collapsed into a single track, from the UCLA/JGI EST project (accession SRX038871). The tracks shown in green represent RNA-Seq coverage, on a log scale, for the locus at three time points (0, 0.5, and 2 h) after Chlamydomonas cells were transferred to nitrogen-depleted medium. The JGI best gene model for the V4 assembly, FM4, and two different gene models predicted by the Augustus algorithm (Au5 and Au10.2), are shown in green and red, respectively. Thick blocks represent exons, and thin blocks represent UTRs, and arrowed lines represent introns. RNA-Seq, 454, and EST coverage were used to inform the manual construction of a single gene model at each locus, labeled user model and shown in blue. The PDAT1 gene is on the left, and the gene model on the right represents an expressed hypothetical protein whose expression is decreased in nitrogen starvation. The blue circles highlight new exons suggested by the patterns of RNA-Seq coverage and incorporated by manual curation into revised gene models. The complete data set is available via the UCSC browser on line.