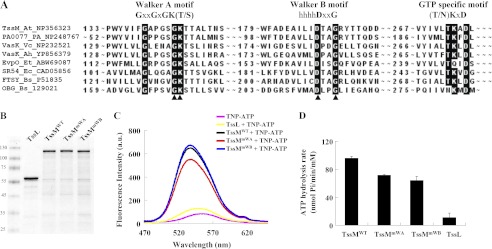
FIGURE 5.
TssM possesses a Walker B motif required for its ATP hydrolysis. A, Walker A, Walker B, and potential GTP-specific motifs were revealed by amino acid sequence alignment of selected T6SS TssM orthologs and other GTPase family members. The conserved amino acid residues of each motif are highlighted; the arrowheads indicate the amino acids targeted for mutagenesis. The proteins used for alignments are indicated in the order of their protein name, organism, and GenbankTM accession number. At, A. tumefaciens C58; PA, P. aeruginosa PAO1; Vc, V. cholerae O1; Ah, A. hydrophila ATCC 7966; Et, E. tarda PPD130/91; Ec, E. coli CT18; Bs, Bacillus subtilis. B, DDM-extracted membrane proteins from E. coli EN2 strain expressing the WT or a variant of TssM-His (TssMWT: pACYC-TssL-TssM-His; TssMmWA: pACYC-TssL-TssMG144A/K145A-His; TssMmWB: and pACYC-TssL-TssMD188A/G191A-His) or TssL-His (TssL: pET-TssL-His) were purified in parallel using Ni-NTA resin. The purity of TssMWT, TssMmWA, TssMmWB, and TssL used for ATP binding and hydrolysis assays was examined by Coomassie Blue-stained SDS-PAGE. The sizes of the molecular mass standards (kDa; Fermentas) are indicated. C, ATP binding activity of TssM and its variants. Fluorescence spectra of TNP-ATP bound to TssM and its variants were detected in the range of 470 to 650 nm. The spectrum for each sample was indicated as WT TssM-His (black, TssMWT +TNP-ATP), TssMG144A/K145A-His (red, TssMmWA +TNP-ATP), TssMD188A/G191A-His (blue, TssMmWB +TNP-ATP), TssL-His (yellow, TssL +TNP-ATP), or TNP-ATP alone (pink). D, ATP hydrolysis activity of TssM and its variants. ATP hydrolysis activity was determined using the malachite green assay to measure the free inorganic phosphate released upon ATP hydrolysis. Average values for ATP hydrolysis activity from three independent experiments are shown with standard deviations.