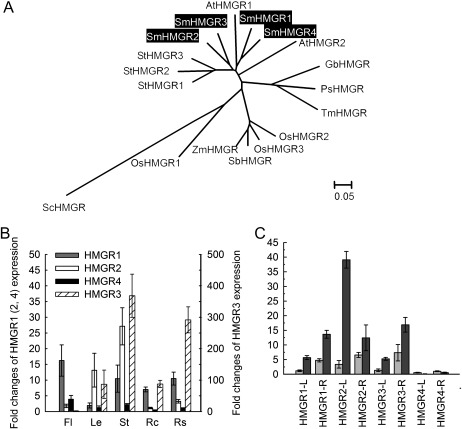
Fig. 3.
Expression patterns of SmHMGR genes and the phylogenetic relationship of their deduced proteins with various other plant species. (A) Phylogenetic relationship of HMGRs in various plant species. The rooted Neighbor–Joining tree was constructed using the MEGA program (version 4.0) with default parameters. ScHMGR (Saccharomyces cerevisiae, AAA34676) was used as an outgroup. HMGRs included are Arabidopsis AtHMGR1 (CAA33139), AtHMGR2 (AAA67317), Solanum tuberosum StHMGR1 (AAA93498), StHMGR2 (AAB52551), StHMGR3 (AAB52552), rice OsHMGR1 (AAA21720), OsHMGR2 (AAD08820), OsHMGR3 (AF110382), Zea mays ZmHMGR (O24594), Sorghum bicolor SbHMGR (XP_002445887), Ginkgo biloba GbHMGR (AAU89123), Picea sitchensis PsHMGR (ACN40476), Taxus×media TmHMGR (AAQ82685), and four S. miltiorrhiza SmHMGRs (highlighted). (B) Fold changes of SmHMGR genes in flowers (Fl), leaves (Le), stems (St), root cortices (Rc), and root steles (Rs) of S. miltiorrhiza plants grown in soil. The expression level of SmHMGR4 in root steles was arbitrarily set to 1. (C) Fold changes of SmHMGRs in leaves (L) and roots (R) of S. miltiorrhiza plantlets treated with MeJA for 0 h and 24 h. The level of SmHMGR4 in roots of plantlets without treatment was arbitrarily set to 1.