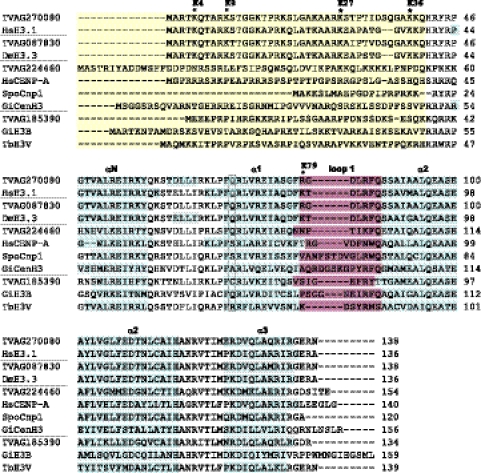
Fig 1.
Analysis of protein sequences of H3 and H3 variants. Sequences are grouped with respect to putative function of the proteins (core histones, H3.3 variant histones, centromeric histones, histone variants with unknown function). Divergent N-terminal extensions (yellow) are not properly aligned between species. Positions of predicted α helices within the histone-fold domain are marked (blue). A conserved glutamine residue of core H3 (boxed) is substituted in all centromeric histones except the histone TVAG_224460. The loop 1 region (purple) shows amino acid insertions in the centromeric histones. Sites of conserved lysine residues known to play a role in gene activation or repression are marked by asterisks; the labels reflect the numbering commonly used for core H3. The TrichDB accession numbers for T. vaginalis sequences are as follows: TVAG_270080, core H3; TVAG_087830, H3.3 variant; TVAG_224460, CenH3; TVAG_185390, H3T (unknown function). NCBI protein database accession numbers are as follows: SpoCnp1, Schizosaccharomyces pombe Cnp1, NP_596473; HsCENP-A, Homo sapiens CENP-A, NP_001800; HsH3.1, Homo sapiens H3.1, NP_003520; GiCenH3, Giardia intestinalis CenH3, EDO81729; GiH3B, Giardia intestinalis H3B, EDO76497; DmH3.3, Drosophila melanogaster H3.3, CAA57080; TbH3V, Trypanosoma brucei H3V, EAN78895.