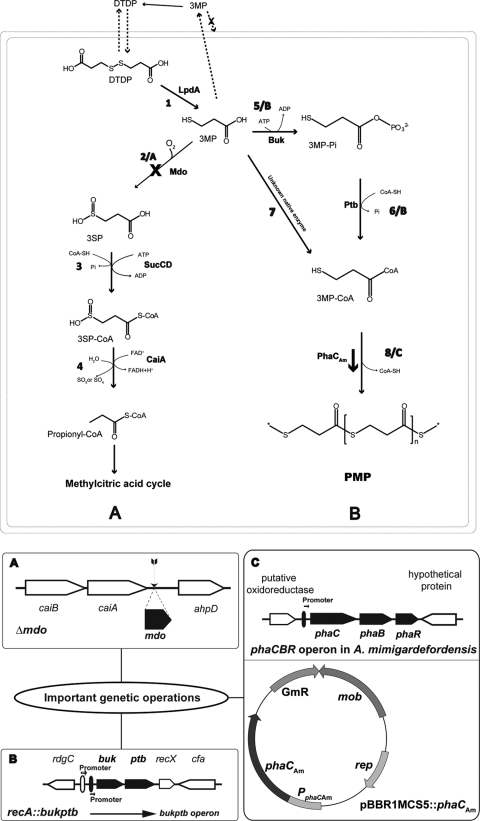
Fig 1.
Predicted DTDP degradation and PMP production pathway. (A) Inherent DTDP degradation pathway of A. mimigardefordensis. (B) DTDP-to-PMP conversion pathway in the engineered strain. The double box indicates the A. mimigardefordensis cell. The deletion of mdo is indicated with a bold X. Dashed arrows across the periplast indicate transmembrane transport of relevant molecules. The bold arrow indicates the overexpression of phaCAm in plasmid pBBR1MCS5:: phaCAm. All reactions catalyzed by enzymes are numbered. Bold A, B, and C indicate three important manipulations for improved PMP production. The detailed genetic operations of these three steps are given at the bottom. The buk-ptb operon originates from C. acetobutylicum and is regulated under its native promoter. Other genes in this figure originate from A. mimigardefordensis DPN7 and are regulated under their own promoters. 3MP, 3-mercaptopropionic acid; DTDP, 3,3′-dithiodipropionic acid; 3SP, 3-sulfinopropionic acid; lpdA, disulfide reductase; mdo, thiol dioxygenase; sucCD, succinyl-CoA synthetase; caiA, acyl-CoA dehydrogenase; buk, butyrate kinase; ptb, phosphotransbutyrylase; phaCAm, poly(hydroxyalkanoic acid) synthase; caiB, acyl-CoA transferase; ahpD, alkylhydroperoxidase; rdgC, recombination-associated protein; recA, recombination protein; recX, recombination regulator protein; cfa, cyclopropane-fatty-acyl-phospholipid synthase; phaB, acetoacetyl-CoA reductase; phaR, polyhydroxyalkanoate synthesis repressor protein; PphaCAm, native promoter of phaCAm.