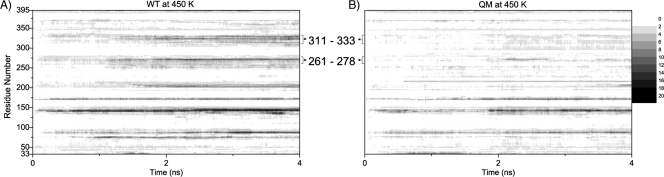
Fig 7.
Contour plots of RMSD (Angstrom) values for the Cα atom of residues (33 to 395) during simulation time (ns) for the wild type (WT) (A) and the quadruple mutant (QM) (B). Graphs are grayscaled from white (0) to black (20) with increments of 2. The regions between amino acids 261 and 278 and between 311 to 333 are labeled. Significant destabilizations of these loops are observed after a 1-ns simulation time for the WT structure at 450 K (dark-shaded regions). On the other hand, the cumulative effect of the given mutations for the QM structure significantly enhances the stabilization of these loops (lighter shades of gray) at 450 K.