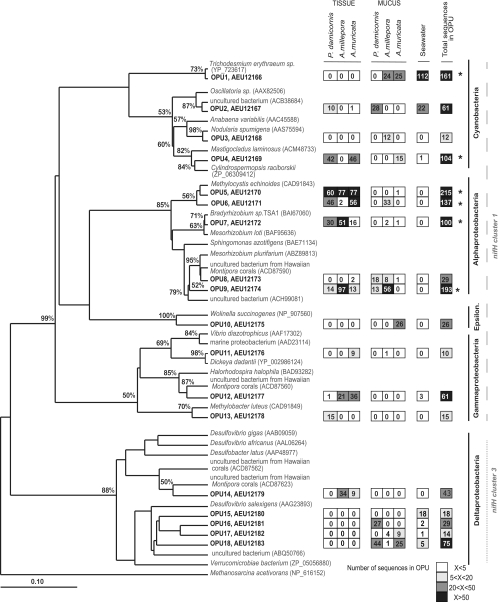
Fig 2.
Phylogeny and composition of the nifH operational protein units (OPU) representing more than 10 sequences (cutoff, 0.1) (n = 1,347 sequences). OPUs from this study are represented in bold with associated protein accession numbers. The maximum-likelihood phylogenetic tree was generated in ARB and includes bootstrap support of 1,000 iterations. The tree was rooted with the nifH protein sequence of an archaeon (Methanosarcina acetivorans). The heat map to the right of the tree represents the total number of sequences retrieved for each OPU for each coral species (mucus and tissue separately) and seawater, with highest values (>50) in black and lowest (<5) in white. OPUs representing over 100 sequences are indicated with asterisks. Sequences from known phylogenies are indicated by species names and protein GenBank accession numbers in parentheses. Classes of bacteria are indicated to the far right of tree (Epsilon. stands for Epsilonproteobacteria), and nifH clusters are indicated with different dashed lines.