Abstract
Elevated levels of DnaA cause excessive initiation, which leads to an increased level of double-strand breaks that are proposed to arise when newly formed replication forks collide from behind with stalled or collapsed forks. These double-strand breaks are toxic in mutants that are unable to repair them. Using a multicopy suppressor assay to identify genes that suppress this toxicity, we isolated a plasmid carrying a gene whose function had been unknown. This gene, carried by the cryptic rac prophage, has been named rcbA for its ability to reduce the frequency of chromosome breaks. Our study shows that the colony formation of strains bearing mutations in rep, recG, and rcbA, like recA and recB mutants, is inhibited by an oversupply of DnaA and that a multicopy plasmid carrying rcbA neutralizes this inhibition. These and other results suggest that rcbA helps to maintain the integrity of the bacterial chromosome by lowering the steady-state level of double-strand breaks.
INTRODUCTION
Chromosomal DNA replication is coordinated to occur once per cell cycle. In Escherichia coli, the initiation of DNA replication requires the recognition of specific sequences in its single replication origin (oriC) by DnaA bound to ATP, which leads to the stepwise assembly of the molecular machinery at each replication fork (reviewed in references 46, 54, and 55). One enzymatic component of this molecular machine is DnaB, the replicative helicase that unwinds the parental duplex DNA as the replication forks advance under a bidirectional mode of fork movement from oriC. Other components are a primase that forms primers on each strand of the parental template DNA for leading- and lagging-strand DNA synthesis and a dimeric DNA polymerase III holoenzyme that copies each parental DNA. Recent evidence indicates that a second DNA helicase, named Rep, interacts with DnaB to facilitate fork movement (36).
Several independent mechanisms control the initiation process so that it occurs only once during each cell cycle (reviewed in references 46, 54, and 55). One mechanism involves SeqA, which specifically recognizes hemimethylated GATC sequences that transiently exist after a new round of DNA replication (59, 71, 73, 93). The specific binding of SeqA to these sequences that are abundant in oriC is thought to sequester the replication origin from DnaA and other replication proteins (6, 71). The second mechanism requires Hda complexed with the β clamp (47, 89). When bound to DNA, this complex stimulates the hydrolysis of ATP bound to DnaA. Because DnaA complexed with ATP is active in initiation, whereas DnaA-ADP is feeble, the interaction of the Hda-β clamp complex regulates the frequency of initiation by affecting the activity of DnaA. The third mechanism relies on a site in the bacterial chromosome named datA (48). On the basis that several hundred DnaA molecules can apparently bind at this locus and that the deletion of this site leads to extra initiations, datA was proposed to titrate DnaA when in excess to avert extra initiations. A separate mechanism also involves the binding of DnaA to other sites in the bacterial chromosome (30, 31, 46). Two sites, named DARS1 and DARS2, for DnaA-reactivating sequence (DARS), contain DnaA box sequences like datA, but the DARS sites stimulate the dissociation of ADP bound to DnaA to permit DnaA to bind to ATP, which is more abundant than ADP in vivo. The effect of DARS1 and DARS2 on the stimulation of the exchange of the nucleotide bound to DnaA is like that of anionic phospholipids (13, 82, 98). The cellular abundance of DnaA also influences the frequency of initiation (38). One process that acts at the level of dnaA expression involves SeqA. Several GATC sequences are in the dnaA promoter region that is bound by SeqA when hemimethylated (20). Hence, SeqA represses dnaA expression during the period shortly after the promoter region has been duplicated but not after the sequences become methylated by DNA adenine methylase. The promoter region also contains DnaA boxes that are recognized by DnaA-ATP to control dnaA expression by autoregulation (3, 17). Moreover, an oversupply of DnaA causes more frequent initiations (4, 56, 84, 85). However, initiation remains synchronous after a mutation of the multiple GATC sites recognized by SeqA in the dnaA promoter (95), suggesting that the inability of SeqA to sequester the promoter does not lead to a sufficiently higher level of DnaA that would promote unscheduled initiation. In contrast, initiation becomes asynchronous due to reinitiation when the corresponding GATC sites in oriC that are recognized by SeqA are inactivated by mutation (7).
Previously, we showed that an increased level of DnaA causes more frequent initiations and an increase in the abundance of double-strand breaks (DSBs) that are toxic in a recA or recB mutant that is defective in DSB repair (84). The level of DSBs presumably increases when the new forks collide from behind with stalled and collapsed replication forks; toxicity follows from the inability to repair them. Whereas surplus DnaA should also repress the expression of the nrdAB-yfaE operon, this has no detectable effect on the cellular abundance of ribonucleotide reductase (33). Hence, the DSBs do not evidently arise from a reduction in both the levels of ribonucleotide reductase and deoxynucleoside triphosphates (dNTPs), which would otherwise lead to stalled forks, followed by a fork collapse. The extra initiations described above also suggest that the oversupply of DnaA surpasses the regulatory pathways that control the frequency of initiation. If so, increasing the copy number of a gene encoding a critical regulatory factor may suppress the lethal effect. To test this idea, we used a multicopy suppressor approach to select for plasmids carrying chromosomal DNA fragments from a plasmid library that suppressed the lethal effect caused by the induced expression of wild-type dnaA (29). We showed genetically that hda; dnaN, which encodes the β clamp of the DNA polymerase III holoenzyme; datA; or seqA in a multicopy plasmid (pACYC184) suppressed the toxic effect caused by elevated dnaA+ expression levels in DSB repair-defective mutants. The results suggest that SeqA, datA, and Hda with the β clamp inhibit initiation under conditions that increase the cellular abundance of DnaA (29), such as DNA damage by methyl methanesulfonate, which leads to blocked replication forks (76). We also isolated a plasmid carrying part of the rac prophage. Here, we characterize a gene named rcbA (née ydaC) because it reduces the frequency of chromosome breaks.
MATERIALS AND METHODS
Bacteriological methods, including the multicopy suppressor assay.
Table 1 lists plasmids and strains that were constructed by P1 transduction essentially as described previously (69). These strains were grown, as indicated, in Luria-Bertani (LB) medium or M9 medium supplemented with 0.4% Casamino Acids, 1% (wt/vol) glucose, or 0.5% (wt/vol) l-arabinose at 37°C. Where appropriate for plasmid maintenance, the growth medium was supplemented with ampicillin (100 μg/ml), chloramphenicol (35 μg/ml), kanamycin (25 μg/ml unless noted otherwise), rifampin (100 μg/ml), and/or tetracycline (15 μg/ml). The frequency of colony formation was measured after overnight incubation at 37°C.
Table 1.
E. coli plasmids and strains
| Plasmid or strain | Description or genotypec | Reference and/or sourcea |
|---|---|---|
| Plasmids | ||
| pACYC184 | Catr Tetr | 23 |
| pBR322 | Tetr Ampr | 14 |
| pING1 | Ampr; paraBADaraC | 44 |
| pDS596 | Ampr; dnaA paraBAD | 43 |
| pLST435 M | Ampr; dnaA(T435M)paraBAD | 84 |
| pKC596 | Amprd; dnaAT7 gp10 promoter | 21 |
| pMMF41 | Catr; hda | 29 |
| pMMF83 | Catr; ralR rcbA ydaQ | This work |
| pMMF83-1 | Catr; ralR rcbA | This work |
| pMMF83-2 | Catr; ralR (ochre) rcbA | This work |
| pMMF83-3 | Catr; ralR (frameshift, ochre, opal) rcbA | This work |
| pMMF83-4 | Catr; ralR ydaQ | This work |
| pMMF85 | Catr; rcbA (CW) insert at BamHI site of pACYC184 | This work |
| pMMF86 | Catr; rcbA (CCW) insert at BamHI site of pACYC184 | This work |
| pMMF-D1 | Catr; rcbA insert at EcoRV site of pACYC184 | This work |
| pMMF-D4 | Catr; rcbA insert at EcoRV site of pACYC184 | This work |
| pMMF-D5 | Catr; rcbA insert at EcoRV site of pACYC184 | This work |
| pMMF-D13 | Catr; rcbA insert at EcoRV site of pACYC184 | This work |
| Strains | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17(rK− mK+) supE44 relA1 lac [F′::Tn10 proAB+lacIqlacZΔM15] | Laboratory stock |
| JJC315 | leu6 his4 argE3 lacY1 galK2 ara14 xyl5 mtl1 tsx33 rpsL31 supE44 hsdR recB268::Tn10 | 67 |
| MC1061 | araD139 Δ(araleu)7697 ΔlacX74 galU galK rpsL hsdR2(rK− mK+) mcrB1 | Laboratory stock |
| SK002 | MC1061 recB268::Tn10 | 29 |
| MV1193 | Δ(lac-proAB) rpsL thi endA sbcB15 hsdR4 Δ(recA-srl)306::Tn10 [F′ traD36 proAB lacIqlacZΔM15] | Laboratory stock |
| HMS174 | (λDE3) recA1 hsdR(rK− mK+) Rifr | Laboratory stock |
| MG1655 | F− λ−rph-1 | Laboratory stock |
| JW1341-1b | Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ− ΔrcbA::kan rph-I Δ(rhaD-rhaB)568 hsdR514 | E. coli Genetic Stock Center; 5 |
| MF1341 | MG1655 ΔrcbA::kan | P1 (JW1341-1) × MG1655→Kanr; this work |
| N3793 | thr-1 araC14 leuB6(Am) Δ(gpt-proA)62 lacY1 tsx33 qsrr-0 glnV44(AS) galK2(Oc) λ−Rac-0 hisG4(Oc) rfbC1 mgl51 rpoS396(Am) rpsL3(Strr) kdgK51 xylA5 mtl-1 recG263(;;kan) argE3(Oc) thi-1 | E. coli Genetic Stock Center |
| JW5604-1 | Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ−rph-I Δrep-729::kan Δ(rhaD-rhaB)568 hsdR514 | E. coli Genetic Stock Center; 5 |
| MF1344 | MG1655 Δrep-729::kan | P1 (JW5604-1) × MG1655→Kanr; this work |
| MF0816 | MG1655 Δ(recA-srl)306::Tn10 | P1 (MV1193) × MG1655→Tetr; this work |
| MF0817 | MG1655 recB268::Tn10 | P1 (SK002) × MG1655→Τetr; this work |
| MF1342 | MF1341 Δ(recA-srl)306::Tn10 | P1 (MV1193) × MF1341→Tetr; this work |
| MF1343 | MF1341 recB268::Tn10 | P1 (SK002) × MF1341→Tetr; this work |
| SS1020 | F− λ−rph-1 zjj202::Tn10 dnaC2 | Steven Sandler |
| MF1345 | MG1655 zjj202::Tn10 dnaC2 | P1 (SS1020) × MG1655→Tetr; this work |
| MF1346 | MF1341 zjj202::Tn10 dnaC2 | P1 (SS1020) × MF1341→Tetr; this work |
For strains, the arrows indicate the method of selection after transduction with the P1 bacteriophage.
The ΔrcbA::kan mutation is identical to the ΔydaC784::kan mutation in JW1341-1.
CW, clockwise; CCW, counterclockwise.
See reference 43.
The multicopy suppressor assay measured the effect of induced dnaA expression on viability. After the coelectroporation of the indicated strains with 50 ng each of pACYC184 or a derivative and a dnaA expression plasmid, dilutions were plated onto LB medium supplemented with ampicillin and chloramphenicol. The medium also contained 0.5% (wt/vol) l-arabinose for strains transformed by pDS596 or pLST435M carrying a dnaA allele under araBAD promoter control (43, 84) or 20 μM isopropyl-β-d-thiogalactopyranoside (IPTG) for those transformed by pKC596 carrying dnaA under the control of a bacteriophage T7 RNA polymerase promoter (21). In assays using E. coli MF1341 (ΔrcbA::kan), the plating medium was also supplemented with 50 μg/ml kanamycin.
Isolation and identification of rcbA as a multicopy suppressor.
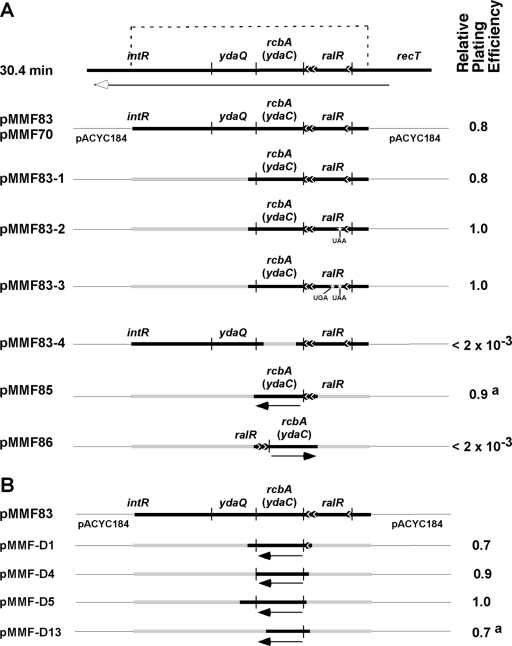
The construction of an E. coli genomic library inserted into pACYC184 was described previously (29). Briefly, the genomic library was coelectroporated with pDS596 into E. coli XL1-Blue (recA1), followed by selection on antibiotic-supplemented LB plates containing 0.5% (wt/vol) l-arabinose to acquire derivatives of pACYC184 that suppressed the toxic effect caused by an elevated level of DnaA. DNA sequence analysis was performed to identify the chromosomal DNA fragment that had been inserted into the BamHI site of pACYC184. Two independently isolated plasmids, named pMMF83 and pMMF70, that contain a 1,301-bp DNA fragment at 30.4 min of the E. coli chromosome were obtained (Fig. 1). The genes in the DNA fragment have the same polarity as the tetracycline resistance gene of pACYC184. To construct pMMF83-1, a 925-bp EcoN1 fragment containing most of the ydaQ gene, a portion of intR, and 247 bp of pACYC184 DNA was removed from pMMF83 by standard recombinant DNA methods. A nonsense mutation in codon 16 of ralR (indicated by boldface type) of primers CACCGTTACTTATGGACAACC and GGTTGTCCATAAGTAACGGTG and a frameshift mutation using primers CATTGTCATATCCATAATTTTTCTC and GAGAAAAATTATGGATATGACAATG, which delete adenine in the second codon of ralR to create nonsense codons for the 17th and 19th amino acids in RalR, were introduced by site-directed mutagenesis with pMMF83-1 as the template DNA according to the manufacturer's instructions (QuikChange; Stratagene). The plasmids constructed were named pMMF83-2 and pMMF83-3, respectively. Primers CAGTCCTCGAGAATTGCATTG and GTTAGTAGGAGTGCCACCTTC, which are complementary to the N- and C-terminal regions of rcbA, respectively, but lack 126 bp of the intervening rcbA coding region from nucleotide 52 to 177, were used to construct pMMF83-4 by reverse PCR amplification (Pfu Turbo; Stratagene), according to the manufacturer's instructions, with pMMF83 as the template DNA. To construct plasmids carrying 98 bp upstream from rcbA, including the indicated promoters in ralR, to 47 bp downstream from rcbA, primers CGTAGTCGACCTGAAATTGACGCCCGATGTTG and CGTAGGATCCTAACGGATGCGAATCCCGAACC, or primers CGTAGGATCCCTGAAATTGACGCCCGATGTTG and CGTAGTCGACTAACGGATGCGAATCCCGAACC, which amplify this region and which also introduce BamHI and SalI sites (indicated by boldface type) near the ends of the amplified DNAs, were designed. After cleavage with the corresponding restriction enzymes, the DNAs were inserted between the BamHI and SalI sites of pACYC184 to create pMMF85 and pMMF86, respectively.
Fig 1.
Deletion analysis and the multicopy suppressor assay identifies rcbA (née ydaC) as a suppressor of lethality caused by overinitiation. The thick and thin horizontal lines represent E. coli DNA and pACYC184, respectively. Vertical lines denote the approximate boundaries of each gene. The shaded thick lines symbolize DNA that has been deleted. The filled circles with arrows represent putative promoters for rcbA, named rcbAp3 at −12 to −28 bp, rcbAp2 at −35 to −63 bp, and rcbAp1 at −151 to −182 bp upstream from rcbA. In the column at the right, the relative plating efficiency was measured by a multicopy suppressor assay with E. coli XL1-Blue (relevant genotype, recA1) cells that were coelectroporated with the dnaA expression plasmid (pDS596) and the indicated plasmid (see Materials and Methods). (A) Genes at 30.4 min of the E. coli chromosome. The line with the arrowhead indicates the polarity of transcription, and the bracket represents the 1,301-bp DNA fragment inserted into the BamHI site of pACYC184 in two independently obtained isolates (pMMF83 and pMMF70). The inserted genes that include part of intR and recT have the same polarity as the tetracycline resistance gene of pACYC184. The construction of other plasmids is described in Materials and Methods. (B) The inserted DNA fragment in pMMF83 was PCR amplified and digested with DNase I in the presence of a manganese ion, and the blunt-end fragments were inserted into the EcoRV site of pACYC184. Fragments inserted into pMMF-D1, pMMF-D4, pMMF-D5, and pMMF-D13 start at 42, 23, 11, and 33 bp upstream of rcbA, respectively, and are oriented with the same polarity as the tetracycline resistance gene of pACYC184. The DNA fragments in pMMF-D1 and pMMF-D5 extend 30 and 60 bp beyond the stop codon of rcbA, respectively. The inserted DNA in pMMF-D4 ends at the stop codon of rcbA, whereas pMMF-D13 lacks 44 bp from the C-terminal coding region of rcbA and is in frame with the remainder of the tetracycline resistance gene. a, small colonies on arabinose-supplemented medium compared with the strain bearing pMMF83.
As an independent method to identify rcbA as the suppressing gene, the DNA fragment inserted into pMMF83 was PCR amplified. After a limited digestion of the amplified DNA (250 ng) for 15 min at room temperature with bovine DNase I (0.2 ng; Sigma) in a volume of 10 μl containing 50 mM Tris-HCl (pH 7.5), 0.1 mg/ml bovine serum albumin (BSA), and 1 mM MnCl2, EDTA was added to a final concentration of 10 mM. The DNA was then purified (QIAprep Spin Miniprep; Qiagen) and end filled with the large fragment of E. coli DNA polymerase I (New England BioLabs) under conditions recommended by the manufacturers to ensure that the ends were blunt. DNA fragments of 100 to 400 bp were purified from an agarose gel (QIAquick gel extraction; Qiagen) and inserted into the EcoRV site of pACYC184 with T4 DNA ligase. The DNA was then purified and coelectroporated with pDS596 into E. coli XL1-Blue. After selecting for transformants on LB plates containing ampicillin, chloramphenicol, and 0.5% l-arabinose, the plasmids obtained (Fig. 1B) were subjected to DNA sequence analysis.
UV irradiation.
A wild-type strain (MC1061 [recB+]) or the isogenic recB mutant (SK002 [recB268::Tn10]) bearing pACYC184, pMMF83, pMMF83-4, or pMMF84 was grown in LB medium at 37°C to a turbidity of between 0.4 and 0.5 optical density (OD) units (595 nm). After the harvested cells were resuspended in 0.1 M MgSO4, the samples were irradiated with UV light (254 nm) at 0.36 J/m2/s. To measure the surviving fraction of cells relative to that of the unirradiated control, bacteria were plated from serial dilutions onto LB plates, which were incubated for 16 h at 37°C. For the plasmid-bearing strains, the liquid and solid media were supplemented with the appropriate antibiotics.
Real-time PCR analysis.
E. coli MG1655, MF1341 (ΔrcbA), or XL1-Blue (recA1) bearing pDS596 and either pACYC184 or a derivative, where indicated, was grown at 37°C in LB medium supplemented with 1% glucose and the appropriate antibiotics to a turbidity of 0.25 OD units (595 nm). After centrifugation, the cells were resuspended in prewarmed LB medium containing either 0.5% (wt/vol) l-arabinose to induce dnaA expression or 1% (wt/vol) glucose and incubated in a shaking water bath at 37°C for 60 min. Real-time PCR analysis was performed with genomic DNA (5 ng), primers (25 pmol each), and SYBR green PCR Master mix (Applied Biosystems) in a 25-μl reaction mixture volume to amplify 100-bp fragments containing oriC or relE, as described previously (29). The default settings of an Applied Biosystems 7500 system and its SDS software package were used to quantify the abundance of these loci relative to a standard curve prepared from genomic DNA isolated from a stationary-phase culture of MG1655. To avoid the problem of interplate variation, which was described previously (29), separate PCR plates were used to determine the abundances of oriC and relE. Their abundances in each DNA sample were quantified in quadruplicate.
Flow cytometry.
The analysis of the DNA content in individual cells was performed essentially as described previously (25, 88). E. coli MG1655 or MF1341 (ΔrcbA) was grown in M9 medium supplemented with 1% (wt/vol) glucose and 0.4% (wt/vol) Casamino Acids to a turbidity of about 0.1 to 0.2 OD units (595 nm). Rifampin and cephalexin were then added to each culture to final concentrations of 100 μg/ml and 10 μg/ml, respectively. After incubation for 3 h at 37°C, the cells were collected by centrifugation, resuspended in a solution containing 10 mM Tris-HCl (pH 7.5) and 10 mM MgSO4 at a volume equal to that of the sample before concentration, and then passed through a 22-gauge needle into 77% ethanol to dilute the samples 10-fold. Prior to analysis, the cells were collected by centrifugation, washed, and then resuspended in a 1/10 volume of the above-described buffer to a concentration of about 2 × 105 cells/ml to 2 × 106 cells/ml. DAPI (4′,6-diamidino-2-phenylindole) staining (2 μg/ml) was done for at least 2 h on ice before flow cytometry.
Pulsed-field gel electrophoresis.
Strains containing the indicated plasmids were grown in LB medium supplemented with 1% (wt/vol) glucose and the appropriate antibiotics at 37°C to a turbidity of 0.25 to 0.3 OD units (595 nm), unless otherwise noted. Where indicated, the cultures were then divided, and l-arabinose was added to one part to a final concentration of 0.5% (wt/vol). After incubation for 1 h at 37°C, the cells were harvested by centrifugation and resuspended in 1/30 of the original culture volume in ice-cold TEE buffer (10 mM Tris-HCl [pH 8.0], 100 mM EDTA, and 10 mM EGTA), and the turbidity (595 nm) was adjusted with this buffer to be essentially identical for each sample. The cells were then incubated for 3 min at 37°C, mixed with an equal volume of prewarmed 2% agarose (SeaPlaque agarose; FMC BioProducts) in TEE buffer, and distributed in 0.25-ml portions into molds. After solidifying at 4°C, the plugs were incubated at 37°C for 2 h in 1.0 ml of TEE buffer containing egg white lysozyme (5 mg/ml; U.S. Biochemicals) and 0.05% (wt/vol) sarcosyl (Sigma), followed by incubation at 52°C for 24 h with proteinase K (1 mg/ml; Invitrogen) and 1% (wt/vol) SDS in TEE buffer. The plugs were washed twice in TEE buffer and either used immediately or stored at 4°C for up to 7 days. Portions (0.05 ml) of each plug were placed into the wells of a 1% agarose gel (Invitrogen Ultra Pure in 45 mM Tris-borate buffer), sealed with 1% agarose (SeaPlaque agarose; FMC BioProducts), and subjected to electrophoresis in 45 mM Tris-borate buffer for 24 h at 150 V, with a switching time ramped from 10 to 120 s with a CHEF-DR II apparatus (Bio-Rad) (92). A mixture of concatemers of λ DNA (lambda ladder PFG marker; New England BioLabs) was used as a molecular weight standard. The gels were stained with ethidium bromide (0.5 μg/ml) for 0.5 h, destained for 1 h, and analyzed with software for a Kodak Image Station 4000R instrument. In addition, essentially identical samples as those used for pulsed-field gel electrophoresis (PFGE) (0.05 ml) were solubilized by the addition to 200 μl of water in a microwave and diluted 40-fold, and the DNA contents of 20-μl portions were determined by ethidium bromide fluorescence (0.5 μg/ml) with a Kodak Image Station 4000R instrument relative to known amounts of DNA, which were used to prepare a standard curve.
RESULTS
The rcbA gene in a plasmid neutralizes the toxic effect caused by overinitiation.
Using a genetic assay that measures the ability of a multicopy plasmid to suppress the toxicity caused by excessive initiation in a recA mutant (see Materials and Methods), we independently obtained two pACYC184 derivatives (pMMF83 and pMMF70) (Fig. 1A) that carry part of the cryptic rac prophage at 30.4 map minutes of the E. coli chromosome. Relying on this multicopy suppressor assay, we then performed a deletion analysis to localize the region with neutralizing activity. After the removal of the segment encoding ydaQ and part of intR in the chromosomal DNA fragment carried in pMMF83, the resulting plasmid (pMMF83-1) remained active in the multicopy suppressor assay, suggesting that rcbA and/or ralR (lar) may be responsible. To test the involvement of ralR, we introduced a stop codon (as in pMMF83-2) and separately constructed a frameshift mutation by deleting a base pair in the ralR gene (pMMF83-3), which then resulted in two nonsense codons for the 17th and 19th amino acids of RalR. The respective plasmids were active in their neutralizing activity, suggesting that ralR is not required. In contrast, the deletion of most of the rcbA gene to form a plasmid named pMMF83-4 abolished the suppressing activity. This deletion also removed a peptide (MREINT) near the center of rcbA and the first 6 amino acids of a hendecapeptide (MQFSRTEVSRN) encoded by the opposite strand near the C terminus of rcbA, so either rcbA, possibly a small RNA (sRNA), or these peptides are required. To confirm the involvement of the DNA fragment carrying rcbA, we removed most of the coding region for ralR, which contains three putative promoters, based on bioinformatics analysis, that are presumed to be involved in rcbA expression (Fig. 1). One promoter is near the start of ralR, whereas the others are near the end of this gene. The plasmids constructed (pMMF85 and pMMF86), which lack the former promoter, varied in their abilities to suppress in a manner dependent on the orientation of rcbA relative to the plasmid. The lack of the upstream promoter in pMMF85 and pMMF86 and the orientation dependence for activity suggest that a promoter in the plasmid is necessary for effective suppression.
To address the involvement of a plasmid promoter and that rcbA, a peptide, or, possibly, an sRNA is required, we isolated the DNA fragment carried in pMMF83 and partially digested it with DNase I, which, in the presence of a manganese ion, cleaves randomly to yield smaller duplex DNAs with blunt ends. Our objective was to identify the smallest chromosomal DNA fragment that conferred activity in the multicopy suppressor assay, which may implicate rcbA or the peptides described above if the DNA is smaller than the rcbA gene or, perhaps, an sRNA. After inserting the collection of DNAs into the EcoRV site within the tetracycline resistance gene of pACYC184, we then transfected the ligation mixture together with the dnaA expression plasmid (pDS596) into a recA mutant. On medium containing antibiotics that selected for both plasmids and l-arabinose that selected for transformants that remained viable despite elevated expression levels of dnaA, we obtained many isolates. Their characterization led to the identification of three plasmids (pMMF-D1, pMMF-D4, and pMMF-D5) carrying the smallest inserted DNAs that are comparable to the parental plasmid (pMMF83) in the multicopy suppressor assay (Fig. 1B). A fourth plasmid (pMMF-D13) was ineffective, as indicated by the smaller colony size upon an oversupply of DnaA, and encodes all but 15 codons at the C-terminal end of rcbA. This plasmid also lacks the first 10 codons of the putative hendecapeptide (MQFSRTEVSRN) encoded near the C terminus of rcbA on the opposite strand. These results suggest that the rcbA gene but not the peptides described above is responsible. If, instead, an sRNA is involved, it may be as long as the rcbA coding region (210 bp). Because point mutations in antisense RNAs have been shown to disrupt base pairing and antisense RNA functions (8, 27, 35, 97) and may also inhibit the activity of an sRNA that interacts with a protein, we have not attempted to introduce nonsense or missense mutations into the rcbA gene. Had we introduced mutations that were inactivating, the results would not exclude the involvement of an sRNA. We provisionally conclude that the function measured in the multicopy suppressor assay is due to rcbA, which may correspond with a protein or sRNA.
For the plasmids shown in Fig. 1B, the polarity of rcbA relative to the plasmid backbone is consistent with the transcription of rcbA from the promoter for the tetracycline resistance gene. Indeed, pMMF-D4 and pMMF-D5 lack the putative promoters in the upstream ralR gene. We also note that rcbA in these plasmids is closer to the promoter for the tetracycline resistance gene, and we speculate that rcbA is expressed at a higher level than that in pMMF85. For pMMF85, the inserted DNA is at the BamHI site that is distal to the promoter for the tetracycline resistance gene and bears two of the three promoters in the upstream ralR gene. This plasmid is ineffective in the multicopy suppressor assay and may express rcbA at a lower level.
The multicopy suppressor assay relies on a plasmid that places dnaA under the control of a regulated promoter. Because the plasmid named pDS596 should require AraC and the catabolite activator protein for the induced transcription of dnaA under araBAD promoter control, we considered that rcbA may act by inhibiting either AraC or the catabolite activator protein or by reducing the intracellular levels of l-arabinose or cyclic AMP. To address these possibilities, we tested whether the rcbA plasmid (pMMF83) could neutralize the lethal effect caused by excess DnaA when its expression was not controlled by the araBAD promoter but by a bacteriophage T7 RNA polymerase promoter. We cotransfected a dnaA expression plasmid (pKC596 bearing dnaA under the control of a T7 RNA polymerase promoter or pDS596 encoding dnaA under araBAD promoter control) with either the empty vector (pACYC184) or the plasmid harboring rcbA (pMMF83) into appropriate strains that are defective in DSB repair and measured growth on antibiotic-supplemented medium in the presence or absence of the inducer. As shown in Table 2, the frequency of colony formation of E. coli HMS174 (relevant genotype, recA1 λDE3) induced to express T7 RNA polymerase or XL1-Blue (recA1) was substantially lower when the level of DnaA was elevated via the respective dnaA plasmid. Compared with the ineffectiveness of pACYC184, the rcbA plasmid maintained the viability of the recA mutants in the presence of the inducer with either dnaA expression plasmid. Plasmids carrying hda, datA, and dnaN were similarly effective (Table 2), confirming previous results (29). Thus, the suppressing effect of these plasmids is not dependent on the expression system. We also examined whether rcbA affects the induced level of DnaA by a quantitative immunoblot analysis of whole-cell lysates. Relative to an internal control of the ribosomal protein L2, the abundances of DnaA encoded by a dnaA plasmid (pDS596) after 1 h of induced expression were comparable in various strains (XL1-Blue [recA1], MG1655, MF1341 [ΔrcbA::kan], or JJC315 [recB268::Tn10]), regardless of whether they carried an rcbA plasmid (pMMF-D4), pACYC184, or no other plasmid (data not shown). These results suggest that the rcbA plasmid does not lead to a lower induced level of DnaA in the multicopy suppressor assay.
Table 2.
An rcbA plasmid suppresses the lethality caused by excess DnaA, regardless of the dnaA expression system, in mutants defective in DSB repaira
| Plasmid | Mean relative plating efficiency ± SD for strain (relevant genotype) and coresident plasmidb |
||
|---|---|---|---|
| XL1-Blue (recA1) with pDS596 (dnaA) | SK002 (recB268::Tn10) with pDS596 (dnaA) | HMS174 (λDE3 recA1) with pKC596 (dnaA) | |
| pACYC184 | <1.1 × 10−3 | <(1.9 ± 1.0) × 10−3 | (2.2 ± 1.1) × 10−3 |
| pMMF83 (rcbA) | 0.9 ± 0.09 | 1.0 ± 0.1 | 0.8 ± 0.15 |
| pMMF41 (hda) | 1.2 | 0.8 | 1.45 ± 0.35 |
| pMMF84 (datA) | 1.1 | 1.0 | 0.7 ± 0.4 |
| pMMF1 (dnaN) | 0.8 | 0.9 | 1.4 ± 0.35 |
The respective strains were coelectroporated with pACYC184, a plasmid encoding rcbA (pMMF83), or derivatives of pACYC184 carrying hda, datA, or dnaN and either pDS596 (50 ng) (dnaA+ under araBAD promoter control) or pKC596 (50 ng) (dnaA+ under the control of a bacteriophage T7 RNA polymerase promoter), as described in Materials and Methods. The strains were then plated onto antibiotic-supplemented LB medium lacking or containing either 0.5% (wt/vol) l-arabinose for strains containing pDS596 or 20 μM IPTG for strains containing pKC596. Incubation was done for 16 h at 37°C. The cotransformation efficiencies for these strains in the absence of an inducer ranged from 8.5 × 104 to 2.3 × 105 transformants per μg of pACYC184 DNA.
The relative plating efficiency is the ratio of the number of colonies observed upon induced dnaA expression divided by the number of colonies in the absence of an inducer. The standard deviation was determined from at least three independent experiments. The shaded values were reported previously (29).
rcbA does not appear to act in DSB repair.
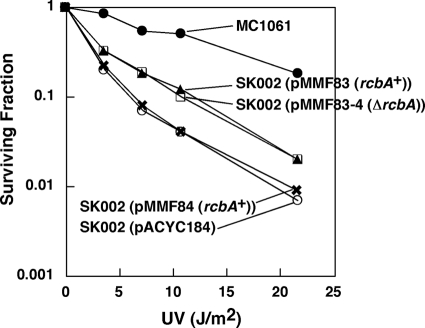
As the rcbA plasmid alleviates the toxic effect of elevated dnaA expression levels in recA and recB mutants (Table 2), a formal possibility is that the rcbA gene product acts in DSB repair by functionally replacing RecA and RecB (reviewed in references 51, 68, and 79). It seems unlikely that the rcbA of 210 bp can encode the multiple activities of RecA or that it can substitute for RecB in the RecBCD complex. Nevertheless, because DSBs appear to arise when replication forks encounter pyrimidine dimers, as suggested by a previous study of UV-treated murine fibroblasts (32), we compared the effects of UV irradiation on a ΔrcbA::kan mutant (MF1341) (see below) to those on an isogenic rcbA+ strain and determined that both strains were comparably UV resistant (data not shown). We also asked whether plasmids carrying rcbA could reduce the UV sensitivity of recA or recB strains. As shown in Fig. 2, plasmids containing or lacking rcbA (pMMF83 and pMMF83-4, respectively) and also carrying the flanking genes slightly increased the UV resistance of the recB mutant, whereas a plasmid (pMMF85) carrying only rcbA was as inactive as the empty vector. Both pMMF83 and pMMF83-4 carry ydaQ and ralR. RalR is thought to protect the bacterial chromosome from degradation by the type 1 restriction-modification system when unmodified DNA is generated by homologous recombination (12). Because the recB mutant carries a mutation in hdsR, which encodes the endonuclease that cleaves unmodified DNA, we can exclude this mechanism as an explanation for the increased UV resistance of the strain carrying plasmids harboring ralR in Fig. 2. Whereas this evidence suggests that ydaQ increases the UV resistance of the recB mutant, none of the plasmids shown in Fig. 2 showed any effect on the survival of a recA mutant after UV irradiation (data not shown). Hence, it is tenuous to attribute a role of ydaQ in the repair of UV-induced lesions.
Fig 2.
An increased gene dosage of rcbA does not confer UV resistance to a recB mutant. E. coli MC1061 (recB+) or SK002 (recB268::Tn10) bearing pACYC184, pMMF83, pMMF83-4, or pMMF85 was grown at 37°C in LB medium to a turbidity of about 0.4 to 0.5 OD units (595 nm) and then irradiated with increasing doses of UV light, as described in Materials and Methods. Bacteria were then plated onto LB plates, followed by incubation at 37°C for 16 h to measure the frequency of colony formation of the treated samples relative to that of the untreated controls. For the plasmid-bearing strains, the medium was supplemented with the appropriate antibiotics.
As with UV irradiation, DSBs arise when cells are treated with methyl methanesulfonate, which alkylates DNA; hydroxyurea, which inhibits ribonucleotide reductase to deplete dNTP pools (28, 58, 77); or zeocin (InvivoGen), which is in a group of compounds related to bleomycin (24). We established that the ΔrcbA::kan mutant (MF1341) was as insensitive to these compounds as an isogenic rcbA+ strain (data not shown). Together, these observations suggest that the rcbA gene product does not substitute for RecA or RecB or act in DSB repair.
DnaA overexpression is lethal in a ΔrcbA strain.
The Keio collection is a set of mutants constructed by the deletion of all nonessential genes of E. coli (5) and includes a ΔrcbA::kan mutant (originally ΔydaC::kan). This evidence indicating that rcbA is not essential contrasts with the profound neutralizing effect of extra copies of rcbA on toxicity caused by excessive initiation that is comparable with those of hda, dnaN, and datA (29). In contrast, with the multicopy suppressor assay, which relies on the effect of an elevated gene dosage, we investigated what happens in the absence of rcbA function by studying a null rcbA mutant. As it is possible that the ΔrcbA mutant in the Keio collection may have acquired other compensatory mutations that may obscure the phenotype caused by the lack of an rcbA function, we transduced the ΔrcbA::kan mutation into a wild-type strain (MG1655). Relative to the titer of the P1 lysate, we obtained kanamycin-resistant transductants at a frequency comparable to that of the transduction of other nonessential E. coli genes, suggesting that the replacement of the wild-type gene with the ΔrcbA::kan mutation does not require a spontaneously arising mutation that suppresses the deficiency of rcbA. However, colonies of the constructed ΔrcbA::kan mutant (MF1341) were about 2-fold smaller than those of the isogenic rcbA+ strain (data not shown). When the mutation was combined with a dnaC2(Ts) mutation (MF1346 [dnaC2 ΔrcbA]), the doubling time of the strain in M9 medium supplemented with 1% (wt/vol) glucose and 0.4% (wt/vol) Casamino Acids at the permissive temperature (30°C) was 79 min, compared with 66 min for the dnaC2 strain (MF1345) (data not shown). Apparently, rcbA improves the growth rate of a wild-type strain and even more so when dnaC function has been compromised.
As shown in Table 3, induced dnaA expression caused a 103-fold reduction in the frequency of colony formation of the ΔrcbA::kan mutant compared with the uninduced control. Compared with the ineffectiveness of the empty vector, plasmids harboring only rcbA (pMMF-D4) or hda (pMMF41) suppressed this lethality. Of interest, the hda plasmid was shown previously to neutralize the toxic effect caused by surplus DnaA in recA or recB mutants (29). With a dnaA allele (T435M) that replaces threonine 435 with methionine and is defective in both the recognition of the DnaA boxes in oriC and initiation (90), its expression did not reduce viability (Table 3). In contrast, the overexpression of dnaA+ did not cause lethality in the isogenic rcbA+ strain (MG1655). Together, these results strongly suggest that overinitiation is toxic in the ΔrcbA::kan mutant.
Table 3.
Plasmids carrying rcbA or hda suppress the lethality caused by an oversupply of DnaA in a ΔrcbA straina
| Strain (relevant genotype) | Mean doubling time (min) ± SD | Cotransformed plasmid | Mean relative plating efficiency ± SDb |
|
|---|---|---|---|---|
| pLST435 M [dnaA(T435 M)] | pDS596 (dnaA+) | |||
| MG1655 (rcbA+) | 23 ± 1.7 | None | 0.9 ± 0.13 | 1.8 ± 0.60 |
| MF1341 (ΔrcbA) | 26 ± 1.7 | None | 0.6 ± 0.24 | <(1.1 ± 0.49) × 10−3 |
| MF1341 (ΔrcbA) | NA | pACYC184 | NA | (6.3 ± 0.42) × 10−3 |
| MF1341 (ΔrcbA) | NA | pMMF-D4 (rcbA) | NA | 1.2 ± 0.11 |
| MF1341 (ΔrcbA) | NA | pMMF41 (hda) | NA | 1.0 ± 0.18 |
The respective strains were grown in LB medium at 37°C to determine the generation time and to prepare electrocompetent cells, which were electroporated with pDS596 or pLST435 M (50 ng) (dnaA allele under araBAD promoter control) and either no second plasmid, pACYC184, or a plasmid carrying rcbA or hda, as described in Materials and Methods and Table 2. Transformants were obtained on LB medium supplemented with 100 μg/ml ampicillin (and 50 μg/ml kanamycin for MF1341), which either lacked or contained 0.5% (wt/vol) l-arabinose. For the strains bearing pACYC184 or its derivatives, the plates also contained 35 μg/ml chloramphenicol. The relative plating efficiency is defined in Table 2. NA, not applicable.
To observe a reduction in the plating efficiency caused by elevated dnaA expression levels in MF1341, kanamycin was required in the plating medium at a concentration of 50 μg/ml.
Of interest, the inviability of the ΔrcbA::kan mutant required kanamycin at 50 μg/ml in the plating medium and was not observed at 25 μg/ml, a concentration at which the strain grew as well as it did without this antibiotic under noninduced conditions (data not shown). Although we do not have an explanation for this, the dependence on this antibiotic for a phenotype was observed previously by Kuzminova et al., who reported that the synthetic lethality of a Δtdk::kan mutation with a recBC(Ts) mutation or of ΔubiH::kan and ΔubiE::kan mutations with recA were observed only in the presence of kanamycin (50). Regardless of the dependence on kanamycin for an observable phenotype, our results indicate that rcbA maintains viability under otherwise lethal conditions.
rcbA does not act at the step of initiation.
Plasmids bearing hda, dnaN, datA, or seqA can counteract the lethal effect of DnaA overproduction (29). The elevated gene dosage apparently reduces the frequency of initiation to a level that the cell can tolerate. These observations raise the possibility that the rcbA plasmid maintains viability under otherwise toxic conditions by reducing the frequency of initiation. If so, the product of rcbA may interfere with the assembly of DnaA and/or other replication proteins at oriC or inhibit the activity of DnaA. To test these ideas, we quantified the abundances of oriC and relE, which is in the terminus region of the chromosome, by real-time PCR analysis. With DNA isolated from log-phase cultures of isogenic rcbA+ and ΔrcbA::kan strains, we then calculated the ratio of these loci to assess the frequency of initiation. As strains lacking hda, seqA, or datA overinitiate, although to various degrees (7, 18, 29, 48, 59, 93), we expected more frequent initiation in the ΔrcbA::kan strain if rcbA is a negative regulator. However, as shown in Fig. 3A, we did not observe increased initiation but instead found that initiation was slightly less frequent in the ΔrcbA mutant than in the isogenic rcbA+ strain. In these strains, the overproduction of DnaA led to comparable increased levels of initiation. In E. coli XL1-Blue, used to select the original rcbA plasmid (Fig. 3B), excess DnaA also led to overinitiation when the strain carried the empty vector or the rcbA plasmid (pMMF-D4) but not when this strain bore a multicopy hda plasmid (pMMF41). These results confirm that Hda negatively regulates the activity of DnaA (47) and strongly suggest that rcbA does not act at the stage of initiation.
Fig 3.
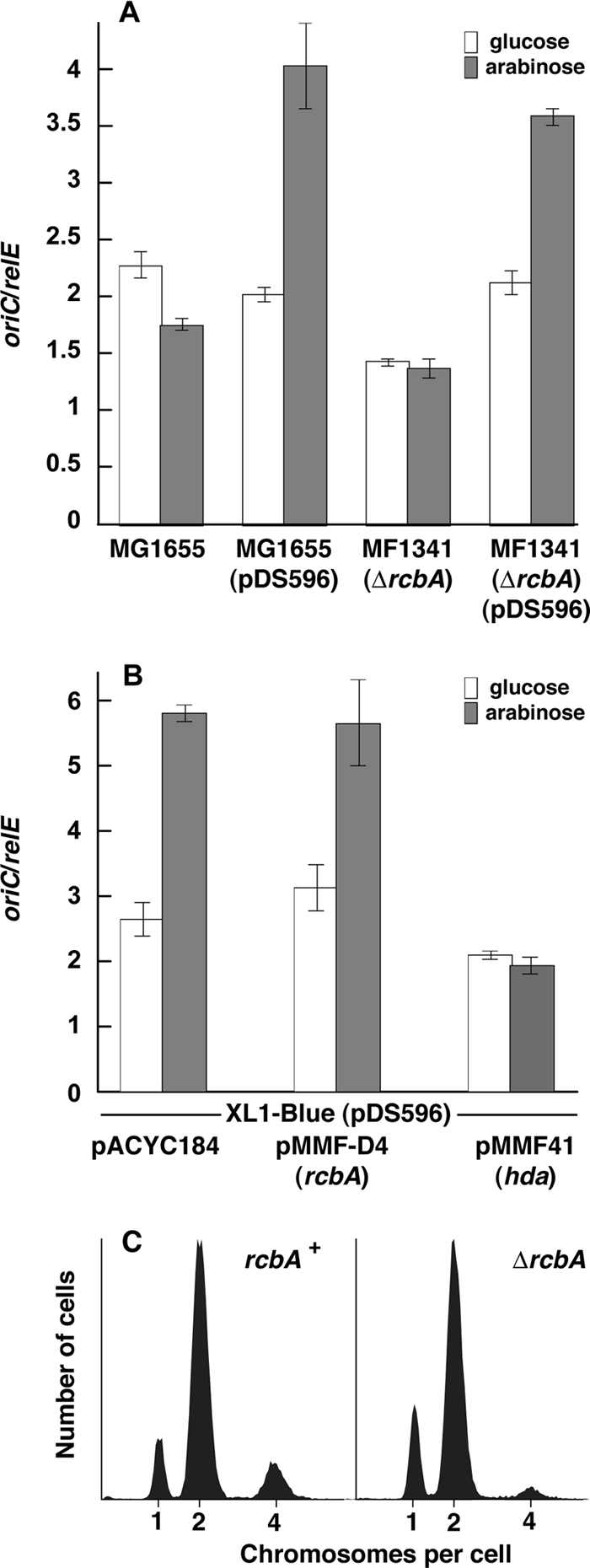
rcbA does not affect the frequency of initiation. (A and B) The frequency of initiation of strains bearing the dnaA expression plasmid (pDS596) and pACYC184 or a derivative harboring rcbA or hda, where indicated, was measured by the ratio of the abundance of oriC to that of relE, a locus in the terminus region. The relative abundance of these loci was determined by real-time PCR analysis as described in Materials and Methods. (C) The content of chromosomes per cell in MG1655 and MF1341 (ΔrcbA) was measured by flow cytometry as described previously (25, 88).
We also measured initiation by an independent method of flow cytometry (Fig. 3C). This technique measures the number of chromosomes per cell after the treatment of a log-phase culture with rifampin and cephalexin, which inhibit new rounds of DNA replication and cell division, respectively (86, 87). As new initiations can occur before chromosomes have finished in a rapidly growing culture, a population of wild-type cells may contain a range from 1, 2, and 4, or 2, 4, and 8, chromosomes, depending on the growth conditions. The relative abundance of these chromosomes reflects the frequency of initiation. Strains lacking hda, datA, or seqA have been found to initiate more frequently when analyzed by this technique (7, 16, 19, 48, 93). Our analysis revealed a higher proportion of cells with one chromosome and a lower proportion of cells with four chromosomes in the ΔrcbA mutant than in an isogenic rcbA+ strain (Fig. 3C). Cells with one, two, or four chromosomes of the rcbA+ and ΔrcbA::kan strains were similar in size (data not shown). These results of less frequent initiation in the rcbA mutant combined with the results described below suggest a positive role for rcbA during cell growth.
The rcbA gene suppresses lethality by reducing the level of double-strand breaks.
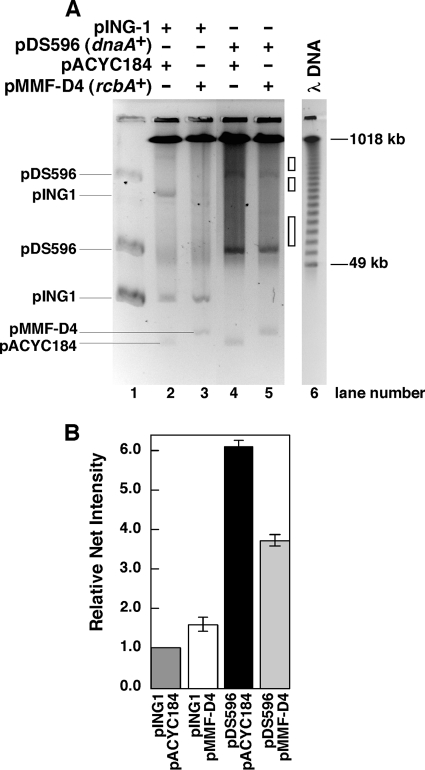
Because overinitiation leads to the accumulation of DSBs that are toxic in strains defective in their repair, the rcbA plasmid may suppress lethality by reducing the level of DSBs. To address this possibility, we measured the level of DSBs in a recB mutant harboring various plasmids by pulsed-field gel electrophoresis (PFGE). As unbroken chromosomes do not enter the gel (11, 67), the linear DNA that migrates into the gel reflects the level of chromosomal fragmentation (50, 68), which is as high as 30 to 40% in strains that hyperinitiate and are unable to repair the resultant DSBs (84). In the experiment shown in Fig. 4, the recB mutant (JJC315) bore plasmids belonging to two sets. A plasmid (pDS596) that carries dnaA under the control of the araBAD promoter or the empty vector (pING1) composed one set. The second set was either a plasmid carrying rcbA (pMMF-D4) or pACYC184. This recB mutant, which Michel et al. used previously to measure DSBs (67), bearing plasmids from both sets was grown to mid-log phase. After the addition of l-arabinose to induce dnaA expression followed by incubation for 1 h, the DNA from comparable numbers of cells was analyzed by PFGE (Fig. 4). We also determined the total amount of sample loaded per lane and quantified the ethidium bromide-stained DNA migrating at the positions marked by the open boxes in Fig. 4A, which excluded the plasmid DNAs. In lanes 2 to 5 of Fig. 4A, the prominent band that migrates at or near the 1,018-kb concatemer of λ DNA (lane 1) presumably corresponds to the 1.8- to 3-Mbp DNA that was observed in this recB mutant (JJC315) (67). Considering the amounts of sample loaded into the respective lanes (lane 3 contains 1.3-fold more sample than lane 2 in Fig. 4A and B), the quantified amounts of fragmented DNA were comparable in the arabinose-treated cells containing the empty vector (pING1) and either pACYC184 or the plasmid carrying rcbA (pMMF-D4). In contrast, in cells bearing the dnaA plasmid (pDS596) and pACYC184, the induced expression of dnaA led to an increase in levels of fragmented DNA. The rcbA plasmid (pMMF-D4), instead of pACYC184, caused about a 1.6-fold reduction in the level of broken DNA (Fig. 4B). Together, these results suggest that additional copies of rcbA reduce the amount of DSBs caused by overinitiation.
Fig 4.
An increased gene dosage of rcbA reduces the abundance of DSBs in a strain defective in DSB repair. (A) JJC315 (recB::Tn10) bearing the indicated plasmids was grown in antibiotic-supplemented LB medium as described in Materials and Methods. After the addition of 0.5% (wt/vol) l-arabinose and incubation for 60 min, the genomic DNA was analyzed by PFGE. Normalized to the amount of DNA loaded in lane 2, which was set at a value of 1, the relative amounts of DNA in lanes 3 to 5 are 1.3, 1.4, and 1.4, respectively. After subtracting the background of fragmented DNA, densitometric analysis revealed that the relative abundances of pDS596 in lanes 4 and 5 were similar and apparently affected little by the rcbA plasmid, with only a 1.1-fold-higher level in the presence of pMMF-D4 than in the presence of pACYC184. (B) The relative abundance of fragmented DNA in each lane represented by the positions of the open boxes in panel A from three replicate experiments was determined to exclude the plasmid DNAs and then normalized relative to the corresponding areas in lane 2, which were set to a value of 1. In this analysis, the respective areas were selected and quantified under conditions in which the densitometric measurements were within the dynamic range of ethidium bromide fluorescence established using increasing known amounts of DNA. The brackets represent the standard deviations of the analysis.
The absence of rcbA leads to increased numbers of double-strand breaks.
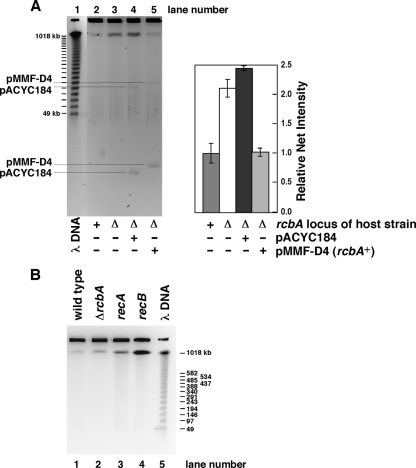
Our observation that increased copy numbers of rcbA lower the DSB level suggests that the absence of rcbA should cause an increase in the amount of DSBs. To test this prediction, we assessed the steady-state level of DSBs in log-phase cultures of a ΔrcbA::kan mutant and an isogenic rcbA+ strain by PFGE and calculated the means and standard deviations from essentially identical samples analyzed in several separate gels (Fig. 5A). Focusing on the DNA migrating at about 1 Mbp, because we could not reliably measure the abundance of smaller broken DNAs by ethidium bromide fluorescence, we observed about a 2-fold-higher level of DSBs in the absence of rcbA function (Fig. 5A). Because the strain is otherwise competent in DSB repair, this increased abundance of DSBs apparently reflects the dynamics of both the production of broken DNA and its repair, which could underestimate the effect of rcbA (see below). Compared with the empty vector (pACYC184) in the ΔrcbA::kan mutant, the rcbA plasmid (pMMF-D4) complemented the null rcbA mutation, as indicated by the decrease in the amount of DSBs to a level comparable with that of the rcbA+ strain.
Fig 5.
The rcbA gene product lowers the steady-state levels of DSBs. (A) As described in Materials and Methods, E. coli strains MG1655 and MF1341 (ΔrcbA::kan) that lacked a plasmid or carried the rcbA plasmid (pMMF-D4) or pACYC184 were grown in LB medium supplemented with 1% glucose and the appropriate antibiotics to a turbidity of 0.2 OD units (595 nm). Bacterial DNA was then analyzed by PFGE. (Left) The amounts of sample analyzed in lanes 3 to 5 (1, 1, and 0.9, respectively) were normalized relative to the amount loaded into lane 2, which was set at a value of 1. (Right) The abundance of the DNA migrating at about 1 Mbp was measured by densitometric analyses of several gels, as described in the legend of Fig. 4. Their amounts are expressed relative to the amount of the 1-Mbp DNA measured in the plasmid-free rcbA+ strain, which was normalized to a value of 1. For the wild type or the isogenic ΔrcbA mutant, the means and standard deviations were calculated from eight samples prepared from three cultures. This analysis also includes the data from lanes 1 and 2 in panel B. For the plasmid-bearing ΔrcbA mutant, the results are from three separate pairs of samples. (B) Bacterial DNA from MG1655, MF1341 (ΔrcbA::kan), MF0816 [Δ(recA-srl)306::Tn10], and MF0817 (recB268::Tn10) was analyzed by PFGE as described in Materials and Methods. Relative to the amount of sample loaded into lane 1, which was normalized to a value of 1, the amounts of sample in lanes 2 to 4 are 0.9, 1.1, and 1.1, respectively.
For comparison, we examined isogenic strains carrying ΔrcbA::kan, Δ(recA-srl)306::Tn10, or recB268::Tn10 mutations (Fig. 5B). Relative to the wild-type strain, we measured about a 2-fold-higher level of the 1-Mbp DNA in the ΔrcbA::kan mutant and have included these data in the summary of results shown in Fig. 5A (right). Visual inspection revealed substantially higher levels of the 1-Mbp DNA in the recA and recB mutants, which substantiates the prominent roles of RecA and RecB in DSB repair (67). Nevertheless, this result of an increased steady-state level of DSBs in the ΔrcbA mutant correlates with its smaller colony size than that of an rcbA+ strain (data not shown), suggesting that the increased numbers of DSBs interfere with cell growth.
Does rcbA act with Rep helicase at the replication fork or with RecG to restart collapsed forks?
On the basis that replication forks appear to move more slowly in a rep mutant than in a wild-type strain (52) and biochemical analyses of Rep (reviewed in references 10, 57, and 60), this DNA helicase is proposed to help replication forks move through regions of the bacterial chromosome that are apparently difficult to copy. One suggestion is that Rep facilitates fork movement through protein-DNA complexes that are believed to impede the progress of the fork (2, 36, 96). Under a separate mechanism that does not exclude the suggestion described above, the apparent involvement of Rep and PriC in reassembling collapsed replication forks may explain the slower-moving forks (39, 42, 78). Rep also physically interacts with DnaB to facilitate DNA unwinding in vitro (2, 36), so Rep and DnaB may coordinate their activities to drive forks forward. Because of these observations, we considered the possibility that the rcbA gene product acts with Rep and DnaB at the fork to reduce stalling. As a rep mutation causes an increased incidence of stalled forks (81), one prediction is that excessive initiation should lead to DSBs in a rep mutant. The additional involvement of Rep in the PriC-dependent pathway of restarting replication forks supports the expectation that the DSBs that fail to reassemble into replication forks should be toxic. We found that an oversupply of DnaA caused a striking reduction in colony size and a slight decrease in the frequency of colony formation in a Δrep::kan mutant (Fig. 6A). If the product of rcbA acts with Rep either to reduce the frequency of stalled forks or to restart collapsed forks, an increased gene dosage of rcbA should not affect the viability of the Δrep::kan mutant when the dnaA expression level is elevated. Our observation that the rcbA plasmid neutralized the toxicity caused by overinitiation suggests that the rcbA gene product does not act with Rep to minimize stalled forks or to restart them (Fig. 6A).
Fig 6.
An rcbA plasmid suppresses the toxic effect caused by the overexpression of dnaA in rep and recG strains. E. coli MF1344 (Δrep::kan) or N3793 (ΔrecG::kan) electroporated with the indicated plasmids was plated onto antibiotic-supplemented LB medium. Where noted, the medium was supplemented with 0.5% (wt/vol) l-arabinose. After incubation for 16 h at 37°C, the ratio of the number of colonies on plates supplemented with l-arabinose to the number on plates without l-arabinose was determined.
RecG is another DNA helicase that may act to rescue forks stalled by lesions in the template DNA and to restart collapsed forks (64–66). This enzyme can interconvert a forked DNA and a Holliday junction, displace R-loops and D-loops, and convert a duplex DNA with a 3′ single-stranded tail into a DNA with a 5′ single-stranded tail. The synthetic lethality of a recG mutation with priA correlates with the ability of RecG together with PriA to rescue a stalled replication fork, suggesting that the physiological role of RecG is to resurrect stalled forks (34, 63). We observed that excess DnaA is inhibitory in a ΔrecG::kan mutant (Fig. 6B). Apparently, RecG is needed to repair the DSBs that accumulate after overinitiation. Using the multicopy suppressor assay, we then tested genetically if the rcbA gene product functions in concert with RecG to avoid generating DSBs. We found that the induced expression of dnaA in the ΔrecG::kan strain caused a reduction in colony size, which was suppressed by the multicopy rcbA plasmid (pMMF-D4). Hence, the product of rcbA appears to act independently of RecG. As an elevated copy number of rcbA suppresses the inhibitory effect caused by surplus DnaA in recG, rep, recA, and recB mutants, the rcbA product does not seem to act with the respective proteins in pathways to resurrect stalled and collapsed replication forks. We consider possible mechanisms for rcbA function below.
DISCUSSION
Here, we describe a role for an uncharacterized gene formerly known as ydaC in minimizing the steady-state level of DSBs. Compared with a wild-type strain, a ΔrcbA mutant has an increased level of DSBs that is remedied by an rcbA plasmid. Moreover, the overproduction of DnaA, which causes excessive initiation that apparently leads to fork collapse when newly formed forks run into stalled forks, is toxic in a ΔrcbA mutant but not in an isogenic rcbA+ strain. We also show that a multicopy plasmid carrying rcbA suppresses the lethality caused by overinitiation in a ΔrcbA::kan mutant and in recA, recB, recG, and rep strains that are defective in DSB repair. In a recB mutant, the rcbA plasmid offsets the increase in numbers of DSBs caused by overinitiation. Together, these results suggest that the product of rcbA reduces the frequency of chromosome breaks.
The rcbA gene is carried by the rac prophage that became integrated into the chromosome of some Gram-negative bacteria over 4.5 million years ago (22, 75). Botstein and Campbell suggested previously that such cryptic prophages may provide a reservoir of potentially useful genes for E. coli (15). In support of this idea, the prophage-carried recE and recT functions, when not repressed in an alternate pathway of homologous recombination (37, 83), and ralR expressed at its endogenous level protect the bacterial chromosome when DNA recombination produces unmodified DNA that would be susceptible to restriction by type 1 restriction-modification systems (12, 26, 49). A recent study showed that prophages increase the resistance to osmotic, oxidative, and acid stresses and also to quinolone and β-lactam antibiotics (94). Our results indicate that a strain lacking rcbA grows more slowly than an isogenic rcbA+ strain, which correlates with the higher steady-state level of DSBs in the absence of rcbA.
Bioinformatics analyses revealed that RcbA is highly conserved among Gram-negative bacteria that carry the rac prophage (our unpublished results). Its abundance of lysine residues and the calculated pI of 10.2 suggest that RcbA binds to DNA in a process that averts DSB formation. Several mechanisms may be considered. One is that the rcbA gene product lowers the frequency of stalled replication forks so that newly formed forks are less likely to collide into them. Alternatively, the rcbA gene product may retard fork movement to lower the risk that newly formed forks will run into stalled forks. The former possibility predicts that the average rate of fork movement is higher, whereas the latter suggests slower-moving forks in an rcbA+ strain than in an rcbA mutant. Alternatively, the product of rcbA may help to restart forks by facilitating the reassembly of the replication fork machinery, which is thought to proceed through two pathways (40, 41, 45, 78, 80). PriC and Rep act in one pathway, but we may exclude this pathway, as the data shown in Fig. 6A suggest that the rcbA gene product does not function with Rep. The second pathway requires the restart proteins PriA, PriB, PriC, and DnaT. If the product of rcbA works in this pathway, an oversupply of DnaA should be toxic in the respective mutants, and an rcbA plasmid should fail to neutralize this toxicity. Presently, we are unable to test these possibilities genetically because the dnaA expression plasmid (pDS596) should require PriA, PriB, and DnaT, and presumably also PriC, for plasmid maintenance (9, 53, 62, 70, 72). To address this obstacle, we plan to construct a plasmid that harbors dnaA but does not require these restart proteins for plasmid maintenance. A separate mechanism is based on the issue that transcription complexes are barriers to replication forks. Amino acid starvation exacerbates this problem, which is relieved by DksA, GreA and GreB, and TraR, transcription factors that minimize the pausing or arrest of RNA polymerase during transcription (1, 61, 74, 91). The rcbA gene product may act in concert with these transcription factors to decrease the frequency of paused transcription complexes. More work is planned to determine the mechanism whereby the product of rcbA reduces DSBs.
ACKNOWLEDGMENTS
We thank Bénédicte Michel (Institut National de la Recherche Agronomique), Steven Sandler (University of Massachusetts, Amherst), and the E. coli Genetic Stock Center for strains. We also thank Jonathan Walton (Michigan State University) for the use of his pulsed-field gel system.
This work was supported by grant RO1 GM090063 from the National Institutes of Health and by the Michigan Agricultural Station.
Footnotes
Published ahead of print 17 February 2012
REFERENCES
- 1. Artsimovitch I, Svetlov V, Anthony L, Burgess RR, Landick R. 2000. RNA polymerases from Bacillus subtilis and Escherichia coli differ in recognition of regulatory signals in vitro. J. Bacteriol. 182:6027–6035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Atkinson J, Gupta MK, McGlynn P. 2011. Interaction of Rep and DnaB on DNA. Nucleic Acids Res. 39:1351–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Atlung T, Clausen ES, Hansen FG. 1985. Autoregulation of the dnaA gene of Escherichia coli K12. Mol. Gen. Genet. 200:442–450 [DOI] [PubMed] [Google Scholar]
- 4. Atlung T, Lobner-Olesen A, Hansen FG. 1987. Overproduction of DnaA protein stimulates initiation of chromosome and minichromosome replication in Escherichia coli. Mol. Gen. Genet. 206:51–59 [DOI] [PubMed] [Google Scholar]
- 5. Baba T, et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bach T, Krekling MA, Skarstad K. 2003. Excess SeqA prolongs sequestration of oriC and delays nucleoid segregation and cell division. EMBO J. 22:315–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bach T, Skarstad K. 2004. Re-replication from non-sequesterable origins generates three-nucleoid cells which divide asymmetrically. Mol. Microbiol. 51:1589–1600 [DOI] [PubMed] [Google Scholar]
- 8. Bardill JP, Zhao X, Hammer BK. 2011. The Vibrio cholerae quorum sensing response is mediated by Hfq-dependent sRNA/mRNA base pairing interactions. Mol. Microbiol. 80:1381–1394 [DOI] [PubMed] [Google Scholar]
- 9. Berges H, Oreglia J, Joseph-Liauzun E, Fayet O. 1997. Isolation and characterization of a priB mutant of Escherichia coli influencing plasmid copy number of delta rop ColE1-type plasmids. J. Bacteriol. 179:956–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bird LE, Subramanya HS, Wigley DB. 1998. Helicases: a unifying structural theme? Curr. Opin. Struct. Biol. 8:14–18 [DOI] [PubMed] [Google Scholar]
- 11. Birren B, Lai E. 1993. Pulse field gel electrophoresis: a practical guide. Academic Press, New York, NY [Google Scholar]
- 12. Blakely GW, Murray NE. 2006. Control of the endonuclease activity of type I restriction-modification systems is required to maintain chromosome integrity following homologous recombination. Mol. Microbiol. 60:883–893 [DOI] [PubMed] [Google Scholar]
- 13. Boeneman K, Crooke E. 2005. Chromosomal replication and the cell membrane. Curr. Opin. Microbiol. 8:143–148 [DOI] [PubMed] [Google Scholar]
- 14. Bolivar F, et al. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95–113 [PubMed] [Google Scholar]
- 15. Botstein D, Campbell A. 1983. Evolution of the lambdoid phages. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 16. Boye E, Stokke T, Kleckner N, Skarstad K. 1996. Coordinating DNA replication initiation with cell growth: differential roles for DnaA and SeqA proteins. Proc. Natl. Acad. Sci. U. S. A. 93:12206–12211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Braun RE, O'Day K, Wright A. 1985. Autoregulation of the DNA replication gene dnaA in E. coli K-12. Cell 40:159–169 [DOI] [PubMed] [Google Scholar]
- 18. Camara JE, et al. 2005. Hda inactivation of DnaA is the predominant mechanism preventing hyperinitiation of Escherichia coli DNA replication. EMBO Rep. 6:736–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Camara JE, Skarstad K, Crooke E. 2003. Controlled initiation of chromosomal replication in Escherichia coli requires functional Hda protein. J. Bacteriol. 185:3244–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Campbell JL, Kleckner N. 1990. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell 62:967–979 [DOI] [PubMed] [Google Scholar]
- 21. Carr KM, Kaguni JM. 1996. The A184V missense mutation of the dnaA5 and dnaA46 alleles confers a defect in ATP binding and thermolability in initiation of Escherichia coli DNA replication. Mol. Microbiol. 20:1307–1318 [DOI] [PubMed] [Google Scholar]
- 22. Casjens S. 2003. Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol. 49:277–300 [DOI] [PubMed] [Google Scholar]
- 23. Chang AC, Cohen SN. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen J, Stubbe J. 2005. Bleomycins: towards better therapeutics. Nat. Rev. Cancer 5:102–112 [DOI] [PubMed] [Google Scholar]
- 25. Chodavarapu S, Gomez R, Vicente M, Kaguni JM. 2008. Escherichia coli Dps interacts with DnaA protein to impede initiation: a model of adaptive mutation. Mol. Microbiol. 67:1331–1346 [DOI] [PubMed] [Google Scholar]
- 26. Clark AJ, Sandler SJ. 1994. Homologous genetic recombination: the pieces begin to fall into place. Crit. Rev. Microbiol. 20:125–142 [DOI] [PubMed] [Google Scholar]
- 27. Colige A, Sokolov BP, Nugent P, Baserga R, Prockop DJ. 1993. Use of an antisense oligonucleotide to inhibit expression of a mutated human procollagen gene (COL1A1) in transfected mouse 3T3 cells. Biochemistry 32:7–11 [DOI] [PubMed] [Google Scholar]
- 28. Davies BW, et al. 2009. Hydroxyurea induces hydroxyl radical-mediated cell death in Escherichia coli. Mol. Cell 36:845–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Felczak MM, Kaguni JM. 2009. DnaAcos hyperinitiates by circumventing regulatory pathways that control the frequency of initiation in Escherichia coli. Mol. Microbiol. 72:1348–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fujimitsu K, Katayama T. 2004. Reactivation of DnaA by DNA sequence-specific nucleotide exchange in vitro. Biochem. Biophys. Res. Commun. 322:411–419 [DOI] [PubMed] [Google Scholar]
- 31. Fujimitsu K, Senriuchi T, Katayama T. 2009. Specific genomic sequences of E. coli promote replicational initiation by directly reactivating ADP-DnaA. Genes Dev. 23:1221–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Garinis GA, et al. 2005. Transcriptome analysis reveals cyclobutane pyrimidine dimers as a major source of UV-induced DNA breaks. EMBO J. 24:3952–3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gon S, et al. 2006. A novel regulatory mechanism couples deoxyribonucleotide synthesis and DNA replication in Escherichia coli. EMBO J. 25:1137–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gregg AV, McGlynn P, Jaktaji RP, Lloyd RG. 2002. Direct rescue of stalled DNA replication forks via the combined action of PriA and RecG helicase activities. Mol. Cell 9:241–251 [DOI] [PubMed] [Google Scholar]
- 35. Guillier M, Gottesman S. 2008. The 5′ end of two redundant sRNAs is involved in the regulation of multiple targets, including their own regulator. Nucleic Acids Res. 36:6781–6794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guy CP, et al. 2009. Rep provides a second motor at the replisome to promote duplication of protein-bound DNA. Mol. Cell 36:654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Handa N, Kobayashi I. 2005. Type III restriction is alleviated by bacteriophage (RecE) homologous recombination function but enhanced by bacterial (RecBCD) function. J. Bacteriol. 187:7362–7373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hansen FG, Christensen BB, Atlung T. 1991. The initiator titration model: computer simulation of chromosome and minichromosome control. Res. Microbiol. 142:161–167 [DOI] [PubMed] [Google Scholar]
- 39. Heller RC, Marians KJ. 2007. Non-replicative helicases at the replication fork. DNA Repair (Amst.) 6:945–952 [DOI] [PubMed] [Google Scholar]
- 40. Heller RC, Marians KJ. 2006. Replisome assembly and the direct restart of stalled replication forks. Nat. Rev. Mol. Cell Biol. 7:932–943 [DOI] [PubMed] [Google Scholar]
- 41. Heller RC, Marians KJ. 2005. The disposition of nascent strands at stalled replication forks dictates the pathway of replisome loading during restart. Mol. Cell 17:733–743 [DOI] [PubMed] [Google Scholar]
- 42. Heller RC, Marians KJ. 2005. Unwinding of the nascent lagging strand by Rep and PriA enables the direct restart of stalled replication forks. J. Biol. Chem. 280:34143–34151 [DOI] [PubMed] [Google Scholar]
- 43. Hwang DS, Kaguni JM. 1988. Purification and characterization of the dnaA46 gene product. J. Biol. Chem. 263:10625–10632 [PubMed] [Google Scholar]
- 44. Johnston S, Lee JH, Ray DS. 1985. High-level expression of M13 gene II protein from an inducible polycistronic messenger RNA. Gene 34:137–145 [DOI] [PubMed] [Google Scholar]
- 45. Jones JM, Nakai H. 1999. Duplex opening by primosome protein PriA for replisome assembly on a recombination intermediate. J. Mol. Biol. 289:503–516 [DOI] [PubMed] [Google Scholar]
- 46. Katayama T, Ozaki S, Keyamura K, Fujimitsu K. 2010. Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat. Rev. Microbiol. 8:163–170 [DOI] [PubMed] [Google Scholar]
- 47. Kato J, Katayama T. 2001. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 20:4253–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kitagawa R, Ozaki T, Moriya S, Ogawa T. 1998. Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 12:3032–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kolodner R, Hall SD, Luisi-DeLuca C. 1994. Homologous pairing proteins encoded by the Escherichia coli recE and recT genes. Mol. Microbiol. 11:23–30 [DOI] [PubMed] [Google Scholar]
- 50. Kouzminova EA, Rotman E, Macomber L, Zhang J, Kuzminov A. 2004. recA-dependent mutants in Escherichia coli reveal strategies to avoid chromosomal fragmentation. Proc. Natl. Acad. Sci. U. S. A. 101:16262–16267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kuzminov A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 63:751–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lane HE, Denhardt DT. 1975. The rep mutation. IV. Slower movement of replication forks in Escherichia coli rep strains. J. Mol. Biol. 97:99–112 [DOI] [PubMed] [Google Scholar]
- 53. Lee EH, Kornberg A. 1991. Replication deficiencies in priA mutants of Escherichia coli lacking the primosomal replication n′ protein. Proc. Natl. Acad. Sci. U. S. A. 88:3029–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Leonard AC, Grimwade JE. 2010. Regulating DnaA complex assembly: it is time to fill the gaps. Curr. Opin. Microbiol. 13:766–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Leonard AC, Grimwade JE. 2011. Regulation of DnaA assembly and activity: taking directions from the genome. Annu. Rev. Microbiol. 65:19–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lobner-Olesen A, Skarstad K, Hansen FG, von Meyenburg K, Boye E. 1989. The DnaA protein determines the initiation mass of Escherichia coli K-12. Cell 57:881–889 [DOI] [PubMed] [Google Scholar]
- 57. Lohman TM, Bjornson KP. 1996. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 65:169–214 [DOI] [PubMed] [Google Scholar]
- 58. Lopes M, et al. 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412:557–561 [DOI] [PubMed] [Google Scholar]
- 59. Lu M, Campbell JL, Boye E, Kleckner N. 1994. SeqA: a negative modulator of replication initiation in E. coli. Cell 77:413–426 [DOI] [PubMed] [Google Scholar]
- 60. Marians KJ. 1997. Helicase structures: a new twist on DNA unwinding. Structure 5:1129–1134 [DOI] [PubMed] [Google Scholar]
- 61. Marr MT, Roberts JW. 2000. Function of transcription cleavage factors GreA and GreB at a regulatory pause site. Mol. Cell 6:1275–1285 [DOI] [PubMed] [Google Scholar]
- 62. Masai H, Arai K. 1989. Escherichia coli dnaT gene function is required for pBR322 plasmid replication but not for R1 plasmid replication. J. Bacteriol. 171:2975–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. McCool JD, Sandler SJ. 2001. Effects of mutations involving cell division, recombination, and chromosome dimer resolution on a priA2::kan mutant. Proc. Natl. Acad. Sci. U. S. A. 98:8203–8210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. McGlynn P, Lloyd RG. 2001. Action of RuvAB at replication fork structures. J. Biol. Chem. 276:41938–41944 [DOI] [PubMed] [Google Scholar]
- 65. McGlynn P, Lloyd RG. 2002. Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell Biol. 3:859–870 [DOI] [PubMed] [Google Scholar]
- 66. McGlynn P, Lloyd RG, Marians KJ. 2001. Formation of Holliday junctions by regression of nascent DNA in intermediates containing stalled replication forks: RecG stimulates regression even when the DNA is negatively supercoiled. Proc. Natl. Acad. Sci. U. S. A. 98:8235–8240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Michel B, Ehrlich SD, Uzest M. 1997. DNA double-strand breaks caused by replication arrest. EMBO J. 16:430–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Michel B, Grompone G, Flores MJ, Bidnenko V. 2004. Multiple pathways process stalled replication forks. Proc. Natl. Acad. Sci. U. S. A. 101:12783–12788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Miller JH. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 70. Minden JS, Marians KJ. 1985. Replication of pBR322 DNA in vitro with purified proteins. Requirement for topoisomerase I in the maintenance of template specificity. J. Biol. Chem. 260:9316–9325 [PubMed] [Google Scholar]
- 71. Nievera C, Torgue JJ, Grimwade JE, Leonard AC. 2006. SeqA blocking of DnaA-oriC interactions ensures staged assembly of the E. coli pre-RC. Mol. Cell 24:581–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nurse P, Zavitz KH, Marians KJ. 1991. Inactivation of the Escherichia coli priA DNA replication protein induces the SOS response. J. Bacteriol. 173:6686–6693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Olsson JA, Nordstrom K, Hjort K, Dasgupta S. 2003. Eclipse-synchrony relationship in Escherichia coli strains with mutations affecting sequestration, initiation of replication and superhelicity of the bacterial chromosome. J. Mol. Biol. 334:919–931 [DOI] [PubMed] [Google Scholar]
- 74. Orlova M, Newlands J, Das A, Goldfarb A, Borukhov S. 1995. Intrinsic transcript cleavage activity of RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 92:4596–4600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Perna NT, et al. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533 [DOI] [PubMed] [Google Scholar]
- 76. Quinones A, Jueterbock WR, Messer W. 1991. DNA lesions that block DNA replication are responsible for the dnaA induction caused by DNA damage. Mol. Gen. Genet. 231:81–87 [DOI] [PubMed] [Google Scholar]
- 77. Salguero I, Guarino E, Guzman EC. 2011. RecA-dependent replication in the nrdA101(Ts) mutant of Escherichia coli under restrictive conditions. J. Bacteriol. 193:2851–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sandler SJ. 2000. Multiple genetic pathways for restarting DNA replication forks in Escherichia coli K-12. Genetics 155:487–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sandler SJ. 2005. Requirements for replication restart proteins during constitutive stable DNA replication in Escherichia coli K-12. Genetics 169:1799–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sandler SJ, McCool JD, Do TT, Johansen RU. 2001. PriA mutations that affect PriA-PriC function during replication restart. Mol. Microbiol. 41:697–704 [DOI] [PubMed] [Google Scholar]
- 81. Seigneur M, Bidnenko V, Ehrlich SD, Michel B. 1998. RuvAB acts at arrested replication forks. Cell 95:419–430 [DOI] [PubMed] [Google Scholar]
- 82. Sekimizu K, Kornberg A. 1988. Cardiolipin activation of dnaA protein, the initiation protein of replication in Escherichia coli. J. Biol. Chem. 263:7131–7135 [PubMed] [Google Scholar]
- 83. Shiraishi K, et al. 2006. The role of UvrD in RecET-mediated illegitimate recombination in Escherichia coli. Genes Genet. Syst. 81:291–297 [DOI] [PubMed] [Google Scholar]
- 84. Simmons LA, Breier AM, Cozzarelli NR, Kaguni JM. 2004. Hyperinitiation of DNA replication in Escherichia coli leads to replication fork collapse and inviability. Mol. Microbiol. 51:349–358 [DOI] [PubMed] [Google Scholar]
- 85. Skarstad K, Lobner-Olesen A, Atlung T, von Meyenburg K, Boye E. 1989. Initiation of DNA replication in Escherichia coli after overproduction of the DnaA protein. Mol. Gen. Genet. 218:50–56 [DOI] [PubMed] [Google Scholar]
- 86. Skarstad K, Steen HB, Boye E. 1983. Cell cycle parameters of slowly growing Escherichia coli B/r studied by flow cytometry. J. Bacteriol. 154:656–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Steen HB, Boye E. 1980. Bacterial growth studied by flow cytometry. Cytometry 1:32–36 [DOI] [PubMed] [Google Scholar]
- 88. Steen HB, Skarstad K, Boye E. 1986. Flow cytometry of bacteria: cell cycle kinetics and effects of antibiotics. Ann. N. Y. Acad. Sci. 468:329–338 [DOI] [PubMed] [Google Scholar]
- 89. Su'etsugu M, Shimuta TR, Ishida T, Kawakami H, Katayama T. 2005. Protein associations in DnaA-ATP hydrolysis mediated by the Hda-replicase clamp complex. J. Biol. Chem. 280:6528–6536 [DOI] [PubMed] [Google Scholar]
- 90. Sutton MD, Kaguni JM. 1997. Threonine 435 of Escherichia coli DnaA protein confers sequence-specific DNA binding activity. J. Biol. Chem. 272:23017–23024 [DOI] [PubMed] [Google Scholar]
- 91. Tehranchi AK, et al. 2010. The transcription factor DksA prevents conflicts between DNA replication and transcription machinery. Cell 141:595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Thoms B, Wackernagel W. 1998. Interaction of RecBCD enzyme with DNA at double-strand breaks produced in UV-irradiated Escherichia coli: requirement for DNA end processing. J. Bacteriol. 180:5639–5645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. von Freiesleben U, Rasmussen KV, Schaechter M. 1994. SeqA limits DnaA activity in replication from oriC in Escherichia coli. Mol. Microbiol. 14:763–772 [DOI] [PubMed] [Google Scholar]
- 94. Wang X, et al. 2010. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 1:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wilkinson TG, II, et al. 2006. The synchrony phenotype persists after elimination of multiple GATC sites from the dnaA promoter of Escherichia coli. J. Bacteriol. 188:4573–4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yancey-Wrona JE, Matson SW. 1992. Bound Lac repressor protein differentially inhibits the unwinding reactions catalyzed by DNA helicases. Nucleic Acids Res. 20:6713–6721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Yu YT, Yuan X, Velicer GJ. 2010. Adaptive evolution of an sRNA that controls Myxococcus development. Science 328:993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Zheng W, Li Z, Skarstad K, Crooke E. 2001. Mutations in DnaA protein suppress the growth arrest of acidic phospholipid-deficient Escherichia coli cells. EMBO J. 20:1164–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]