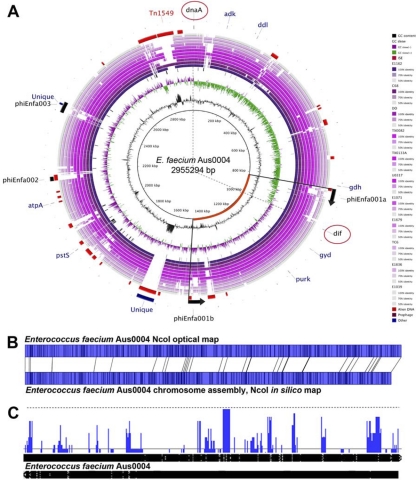
Fig 1.
Analysis of the Enterococcus faecium AUS0004 complete genome. (A) Circular map of AUS0004 by comparative BLASTN analysis against the contigs from the partially assembled genomes of 11 E. faecium strains, showing the locations of a large chromosomal inversion and Aus0004 accessory genome elements. Track identification, moving outwards, is as follows: G+C content, GC skew (G-C/G+C), IS elements, E. faecium isolates (next 11 tracks, as listed here and in Table 1), followed by regions (red arcs) potentially acquired by HGT revealed by AlienHunter, location of prophage, Aus004 unique regions revealed by read mapping against 23 E. faecium genomes (blue arcs), and finally the location of the eight MLST genes. Dotted lines indicate likely replication origin (dnaA) and terminus (dif) and highlight a replichore imbalance, caused by a phiEnfa001-mediated chromosomal inversion. (B) NcoI optical map of E. faecium AUS0004 compared with in silico-derived NcoI map demonstrating correct chromosome assembly. (C) Artemis linear view of Aus0004 chromosome and plasmids (appended), with vertical blue bars identifying the positions of accessory genome elements as determined by read mapping against 23 publicly available partially assembled genome sequences. Increasing height of vertical blue lines on this map indicates increasing specificity for Aus0004.